Abstract
Background:
Congenital heart disease (CHD) is the most widespread congenital disease in newborn babies and is one of the main causes of death worldwide. The causal agent of heart congenital diseases is unknown but genetic factors have an important role in prevalence of disease.Objectives:
The main objective of this research is comparison of the gene expression level of three Gata4, Tbx5 and Nkx2.5 genes in three groups of children between 6 months and 13 year old with congenital heart disease.Patients and Methods:
In this case-control study, 30 samples from each cyanotic and acyanotic patients and 30 samples from healthy children as control were used. RNA extraction was done using commercial kit and gene expression analysis was performed by qRT-PCR approach in three replication using Gata4, Tbx5 and Nkx2.5 genes. Data analysis was done by REST software.Results:
The results of RNA extraction and cDNA synthesis of all sample showed high quantity and quality of genetic materials. Expression level of tested genes was reduced in two patients group. In cyanotic group reduction was more than acyanotic samples. All tested gene were reduced in both group. Tbx5 gene was suppressed more than other genes.Conclusions:
Based on our results we could conclude that a gene family play an important role in cardiogenesis process and heart formation. These genes are closely related together. So a genetic consultation for such diseases on parents of these patients to determine the probable genetic mutations is recommended.Keywords
Heart Disease Gata4 Tbx5 Nkx2.5 Real-time Polymerase Chain Reaction
1. Background
Congenital heart disease (CHD) is the most widespread congenital disease in newborn babies and is one of the main causes of death worldwide [1, 2]. CHD are principal causes of death during the first year of life. They occur in about 5 - 8 of 1000 live births [3, 4]. CHD are caused by defects or malformations in one or more structures of the heart or blood vessels that occur during the 3 - 8 weeks of the first trimester of pregnancy when the heart is being formed [5, 6]. In newborn babies patient the risk of effects and death increases due to delay recognition and referring to medical centers [5]. The main cause of congenital heart diseases in children is unknown but genetic factors have an important role in prevalence of disease [6]. A genetic mutations set have been discovered in the affected families by CHD, that lead to cardiac development defects [7]. When a baby with a septal defect in ventricles and atria is born, he/she has a septal defect in heart that is a type of CHD [8]. The causes of cardiac development defects are unknown but genetic studies have discovered many genes cluster family involving heart development. In addition, the congenital defects of heart can caused by monogenic disorders [9]. Vertebrate heart formation is controlled by the cooperation of transcription factors, which regulate gene expression involved in cardiac development process [10]. A set of genes such as Gata4, Nkx2-5 and Tbx5 are involved at cardiac development from the beginning of the birth. Intra embryonic expression of Gata4 RNA was first revealed in tissue of mouse embryo (7.0 - 7.5 days postcoitum ). Gata4 gene plays an important role in formation of heart-vessel canal and right ventricle [11]. The zinc finger transcription factor Gata4 has been implicated in heart development based on its early expression [12]. The Gata transcription factors play an essential role in precardiogenic splanchnic mesoderm during development and function in retention of cell distinction in adult tissues [13]. Tbx5 transcription factors belongs to the T-box family located on chromosome 12 [14]. T-box 5 protein is product of Tbx5 gene plays an important role in cell differentiation and the formation of tissues and organs during embryonic development. This protein as a transcription factor regulates the activity of other genes by DNA binding action to specific regions of DNA [15]. Csx/Nkx2.5 gene coding an homeobox-containing transcription factor, is a vertebrate homeobox gene with a sequence homology to the Drosophila tinman, which is required for the dorsal mesoderm specification and heart formation and development. Recently, heterozygous mutations of this gene were found to cause the human congenital heart disease. Mutations in this gene cause atrial septal defect with atrioventricular conduction defect, and also Tetralogy of Fallot, which are both heart malformation diseases [16, 17].
2. Objectives
The main objective of this research is comparison of the gene expression level of three Gata4, Tbx5 and Nkx2.5 genes in three groups of children between 6 months and 13 year old with congenital heart disease.
3. Patients and Methods
3.1. Patient Recruitment
This case-control study, was performed in institute of biotechnology of Zabol University.
Required sample size was determined using statistical equation formula accordance with α= 0.05, β = 0.1 [18].
Blood samples was collected from patient referred to Imam Ali hospital of Zahedan. Patients were divided to three principal groups including: the individuals affected by cyanotic congenital heart disease, the individuals affected by acyanotic congenital heart disease and healthy persons as control. In each group, samples were divided to four sub-group, based on theirs age (A, 6 months-1 year; B, 1 - 4 years; C, 4 - 7 years and D, 7 - 13 years). All blood from healthy children were pooled and then served as control. Three milliliter of harvested blood was transferred to 15 mL falcon tubes and stored at -80°C for further use. Patients who were affected by other diseases except CHD diseases such as renal lung, digestive diseases, metabolic, endocrine, chronic infectious diseases, connective tissue and nervous-muscular diseases were deleted.
3.2. RNA Isolation and First Strand Synthesis
Total cellular RNA was isolated from fresh and frozen babies’ blood of 6 months till 13 years old to by RNeasy mini kit (Qiagen, USA). Total RNA was quantified using a Scandrop spectrophotometer (AnalytikaGena, Germany) and RNA quality was assessed by 1.5% agarose gel electrophoresis stained by ethidium bromide and photography by Geldocument (Vilber, France). First-strand cDNA synthesis was carried out in 30 µL of reaction mixture containing 1 µg of total RNA, 6 µL of random hexamers (50 M), 6 µL of 100 mM dithiothreitol (DTT), and 600 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (Qiagene).
3.3. Real-time Polymerase Chain Reaction ( RT-PCR)
Expression analysis of three genes involved in cardinogenesis process was performed using real-time PCR (RG3000, Corbett research). The sequences of used primers are summarized in Table 1. For real-time PCR, 2 µL of the reverse-transcribed cDNA template (50 ng/µL) was added to a final volume of 20 µL of reaction buffer containing 1.5 mM MgCl2, 50 mM KCl, 0.2 mM deoxynucleoside triphosphates, 15 pM each primer and 0.5 U of Taq polymerase in 3 µL of Master Taq polymerase enhancer (Eppendorf). The real time PCR was done on the two biological and three technical replicates. Reaction condition for thermal cycling was 56 for 2 minutes, 95 for 5 minutes followed 40 cycles; 95 for 15 seconds and 65 for 1 minute. A cycle threshold [19] reveals the cycle number at which DNA amplification is detected. The quantity of glyceraldehyde-3-phosphate dehydrogenase( GAPDH )gene was used as internal control [20]. All data were analyzed using REST software and also the Ct values of target genes were normalized to endogenous reference and then were calibrated by 2–ΔΔC [21].
Primers Sequences Used in This Study
| Gene | Primer Name | Forward (5’-3’) | Reverse (5’-3’) | Reference |
|---|---|---|---|---|
| NKx2.5 | Nkx2.5-m-exp-fr/r3 | CTCCGATCCATCCCACTTTA | AGTGTGGAATAGTCGAAAG | Schlesinger et al., 2011 |
| Gata4 | Gata4-m-exp-fr/r2 | CTCCTTACTCCAGCCCCTACC | GCCCCACAATTGACACACACTC | Riazi et al., 2005 |
| Tbx5 | Tbx5-rt-m-f/r1 | TCCGGCTTTCCTGCTAAGA | GGCCAAAGCCCTCATCTGTAT | Schlesinger et al., 2011et al., |
| GAPDH | Reference control | TCCACCACCCTGTTGCTGTAG | GACCACAGTCCATGCCATCATC | Hu et al., 2010 |
4. Results
In this research, we analyzed gene expression level of three Nkx2-5, Tbx5 and Gata4 genes in two cyanotic and acyanothic groups which each group was divided to four sub-groups consist different age patient from 1 month old to 13 years old. These genes are involved in cardiogenesi process (Figure 1).
Interaction of Transcription Factors to Expresses Downstream Gene Regulation
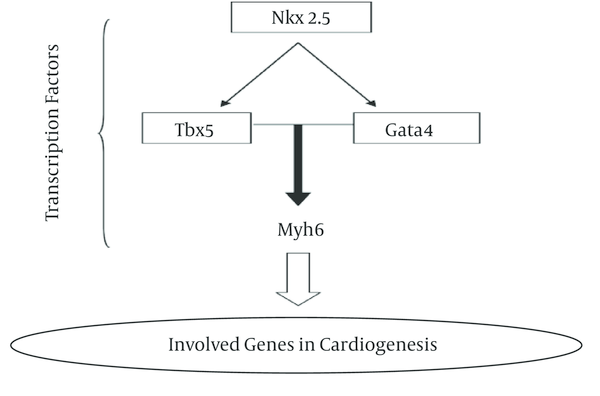
The sequences of all tested genes were choice from literature reviews and then the best primer with low C+G percent was synthesis (Table 1).
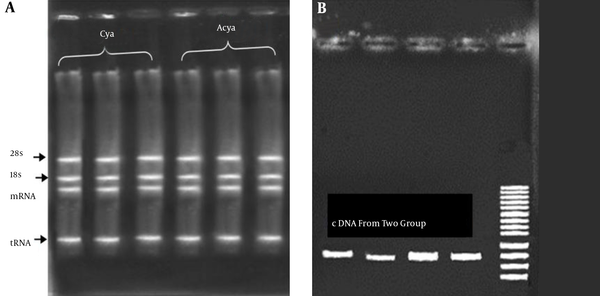
The extraction of RNA was done using fresh blood of patients. The results of RNA extraction of all samples showed a high quantity and quality of genetic materials. An average of 1500 ng/μL of RNA (with OD260/280 = 1.9 to 2) was obtained (Figure 2A). The first strand of cDNA synthesis was performed using commercial kit .The result of cDNA synthesis showed a high quality and quantity of cDNA molecules. To ensure that cDNA was perfectly synthesized a PCR amplification was carried with designed primers (Figure 2B).
RNA Extraction
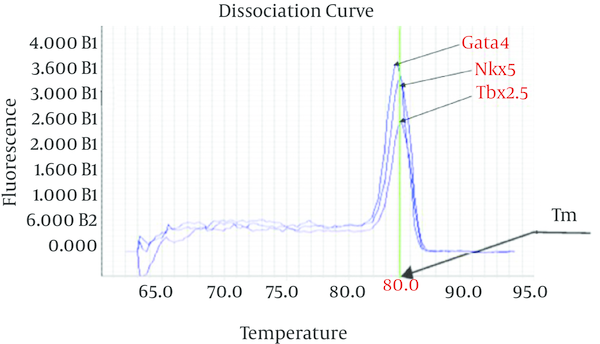
The thermal melting curve of double-stranded DNA related to all used primers in this study was assay during qPCR process. The result of Tm assay showed sharp reduction in the fluorescence and a single and clear melting temperature (Tm) curve with narrow peaks signal (Figure 3).
Dissociation Curves Gata4, Tbx2.5 and Nkx5 Gene of Amplified Cycles in qRT-PCR
Data analysis of gene expression showed that there is not significant different on expression pattern between samples of boys and girls especially in young age children. These results indicate that sexuality has not important role in defect in early cardiogenesis process.
The expression level of three tested genes in two assayed groups was decreased but in cyanotic group was more than acyanotic group (Figures 4 and 5).
Gene Expression Analysis of Cyanotic Patient in Different Age Stages Accordance With the Mean ± SE From Triplicate Sample
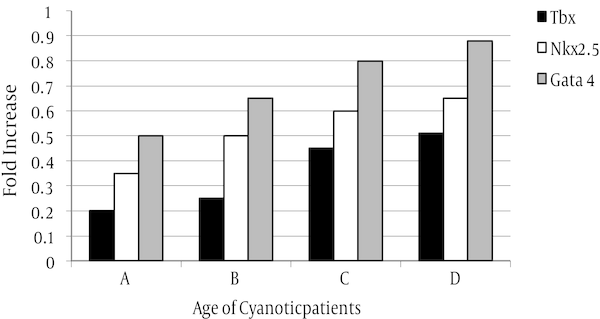
The rate of expression was varied in sub-groups. In both assayed group, expression level of Nkx2.5 gene was reduced in all samples in compared to control whereas, Gata4 gene expression was increased in all sampled when compared with control.
Gene Expression Analysis of Acyanotic Patient in Different Age Stage Accordance With the Mean ± SE From Triplicate Sample
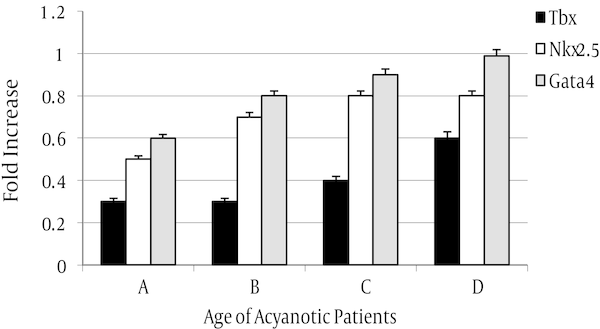
5. Discussion
The results of RNA extraction of all samples showed a high quantity and quality of genetic materials. An average of 1500 ng/μL of RNA was obtained. High quality and quantity of cDNA molecules observed from the analysis. The result of Tm assay showed sharp reduction in the fluorescence and a single and clear melting temperature curve with narrow peaks signal. The analysis showed that there is not significant different on expression pattern between samples of boys and girls especially in young age children. In both assayed group, expression level of Nkx2.5 gene was reduced in all samples in compared to control whereas, Gata4 gene expression was increased in all sampled when compared with control. The expression level of three tested genes in two assayed groups was decreased but in cyanotic group was more than acyanotic group. Heart is one of the first organs which is formed in embryonic period and all the events in the life of organisms is depend on heart’s ability to provide all the needs especially oxygen and nutrients. The defective in heart formation is one of the commonest disorders especially at the time of birth [22]. Three members of the TBx5, Nkx2.5 and Gata family of transcription factors are expressed in the developing heart. The mechanisms underlying this transcription factors specificity are not fully understood but may involve interaction with other tissue-restricted or ubiquitous co-factors [12]. Gene expression analysis in three experimented groups did not show a high significant expression level compared to control patients. Using one Nkx2.5 mutation positive child showed that gene expression of Nkx2.5 transcription factor was significantly reduced than other child in same family [23]. These results were in accordance to our observations in this study. Nkx2.5 gene is the first transcription factor involved in cardiogenesis procedure and its suppression led to reduce of other transcription factors in cardiogenesis complex pathway. The gene encoding Gata4, a zinc finger transcription factor implicated in cardiac gene expression and development, localizes to chromosome region 8p23.1 [24].
The results of this work showed a reduction level of Gata4 gene that this could be due to malfunction of up-stream Nkx2.5 gene. This finding was agree to previous study that have been demonstrated the role of Gata4 and Tbx5 gene to active downstream genes in cardiogenesis [25]. Gata4 function is also relevant for development of heart tissue systems normally [26]. Our observation also showed that in D sub-group (adolescents) in both cyanotic and acyanotic the expression level of Gata4 and Tbx5 genes was low than newborn but this decrease was significantly more than control pool. The gene expression analysis revealed that all tested genes were suppressed in both groups but this reduction was significant in cyanotic groups. Expression analysis of selected samples of girls and boys did not different expression pattern and this indicate that this deification could not related to sexuality. Tbx5 and Gata4 genes are near each other in chromosomal situation and their expression is closely related to each other [25]. Our results suggest that Tbx5 and Gata4 could collaborate to provoke cardiogenesis pathway. The previous studies related to Nkx2.5 and Gata4 mutation showed that the mutant has cardiogenic deficiency [23, 27]. So our observations are according to previous observations and determine the basic role of these two genes in production of structural protein of heart tissue. According to the results we recommended a perfect genetic study on parents of these patients to determine the probable genetic mutations that may be transferred to next generation from parents. Also a genetic consultation for such diseases is recommended. In a complete study, providing chips of carrier’s gene that are involved in cardigenesis pathway in a specific period (age period) in all samples is recommended.
Acknowledgements
References
-
1.
Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal Biochem. 2002;303(1):95-8. [PubMed ID: 11906156]. https://doi.org/10.1006/abio.2001.5564.
-
2.
Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999-2001. Birth Defects Res A Clin Mol Teratol. 2006;76(11):747-56. [PubMed ID: 17051527]. https://doi.org/10.1002/bdra.20294.
-
3.
Shahramian I, Noori NM, Hashemi M, Sharafi E, Baghbanian A. A study of serum levels of leptin, ghrelin and tumour necrosis factor-alpha in child patients with cyanotic and acyanotic, congenital heart disease. JPMA. 2013;63:1332-7.
-
4.
Noori NM, Sadeghi S, Shahramian I, Keshavarz K. Urine beta 2-Microglobolin in the Patients with Congenital Heart Disease. Int Cardiovasc Res J. 2013;7(2):62-6. [PubMed ID: 24757623].
-
5.
Kuehl KS, Loffredo CA, Ferencz C. Failure to diagnose congenital heart disease in infancy. Pediatrics. 1999;103(4 Pt 1):743-7. [PubMed ID: 10103296].
-
6.
Sorrell VL, Pancyzk E, Alpert JS. A new disease: bicuspid aortic valve aortopathy syndrome. Am J Med. 2012;125(4):322-3. [PubMed ID: 22444095]. https://doi.org/10.1016/j.amjmed.2011.11.003.
-
7.
Anderson RH, Wessels A, Vettukattil JJ. Morphology and morphogenesis of atrioventricular septal defect with common atrioventricular junction. World J Pediatr Congenit Heart Surg. 2010;1(1):59-67. [PubMed ID: 23804724]. https://doi.org/10.1177/2150135109360813.
-
8.
Srivastava D, Olson EN. A genetic blueprint for cardiac development. Nature. 2000;407(6801):221-6. [PubMed ID: 11001064]. https://doi.org/10.1038/35025190.
-
9.
Ermis P, Franklin W, Kim J, Moodie D, Parekh D. Electrophysiology procedures in adults with congenital heart disease. Congenit Heart Dis. 2012;7(4):344-8. [PubMed ID: 22536993]. https://doi.org/10.1111/j.1747-0803.2012.00658.x.
-
10.
Hatcher CJ, McDermott DA. Using the TBX5 transcription factor to grow and sculpt the heart. Am J Med Genet A. 2006;140(13):1414-8. [PubMed ID: 16691575]. https://doi.org/10.1002/ajmg.a.31256.
-
11.
Heikinheimo M, Scandrett JM, Wilson DB. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol. 1994;164(2):361-73. [PubMed ID: 8045339]. https://doi.org/10.1006/dbio.1994.1206.
-
12.
Charron F, Nemer M. GATA transcription factors and cardiac development. Semin Cell Dev Biol. 1999;10(1):85-91. [PubMed ID: 10355032]. https://doi.org/10.1006/scdb.1998.0281.
-
13.
Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275(50):38949-52. [PubMed ID: 11042222]. https://doi.org/10.1074/jbc.R000029200.
-
14.
Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life. 2010;62(2):92-102. [PubMed ID: 19960541]. https://doi.org/10.1002/iub.275.
-
15.
Debeer P, Race V, Gewillig M, Devriendt K, Frijns JP. Novel TBX5 mutations in patients with Holt-Oram syndrome. Clin Orthop Relat Res. 2007;462:20-6. [PubMed ID: 17534187]. https://doi.org/10.1097/BLO.0b013e3181123ffe.
-
16.
Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, et al. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovasc Genet. 2013;6(3):238-47. [PubMed ID: 23661673]. https://doi.org/10.1161/CIRCGENETICS.113.000057.
-
17.
Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126(6):1269-80. [PubMed ID: 10021345].
-
18.
Bentler PM, Stein JA. Structural equation models in medical research. Stat Methods Med Res. 1992;1(2):159-81. [PubMed ID: 1341656].
-
19.
Prime AG, Conrath U, Beckers GJ, Flors V, Garcia-Agustin P, Jakab G, et al. Priming: getting ready for battle. Mol Plant Microbe Interact. 2006;19(10):1062-71. [PubMed ID: 17022170]. https://doi.org/10.1094/MPMI-19-1062.
-
20.
Hu B, Wu YM, Wu Z, Phan SH. Nkx2.5/Csx represses myofibroblast differentiation. Am J Respir Cell Mol Biol. 2010;42(2):218-26. [PubMed ID: 19395679]. https://doi.org/10.1165/rcmb.2008-0404OC.
-
21.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402-8. [PubMed ID: 11846609]. https://doi.org/10.1006/meth.2001.1262.
-
22.
Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451(7181):943-8. [PubMed ID: 18288184]. https://doi.org/10.1038/nature06801.
-
23.
Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, et al. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41(11):2072-6. [PubMed ID: 12798584].
-
24.
Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, et al. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet. 1999;83(3):201-6. [PubMed ID: 10096597].
-
25.
Perrino C, Rockman HA. GATA4 and the two sides of gene expression reprogramming. Circ Res. 2006;98(6):715-6. [PubMed ID: 16574910]. https://doi.org/10.1161/01.RES.0000217593.07196.af.
-
26.
Thurisch B, Liang SY, Sarioglu N, Schomburg L, Bungert J, Dame C. Transgenic mice expressing small interfering RNA against Gata4 point to a crucial role of Gata4 in the heart and gonads. J Mol Endocrinol. 2009;43(4):157-69. [PubMed ID: 19491195]. https://doi.org/10.1677/JME-09-0030.
-
27.
Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443-7. [PubMed ID: 12845333]. https://doi.org/10.1038/nature01827.
