Abstract
Background:
E. coli is a common cause of urinary tract infection and bacteremia in humans. That is frequently resistant to β-lactam antibiotics.Objectives:
The aim of this study was to determine the antibiotic resistance and CTX-M extended-spectrum β-lactamases (ESBL) carrying E. coli isolated in hospitals from two different areas in Iran.Methods:
In this descriptive cross-sectional study, 93 clinical isolates of E. coli were collected from urine culture of inpatients in Mashhad and Qom hospitals from August 2013 to February 2014. Bacteria were re-confirmed by Microgen kit. Antimicrobial and ESBL tests were performed using by standard disc diffusion method. Polymerase chain reaction (PCR) was used for detection of plasmid mediated CTX-M-1 and CTX-M-2 ESBLs. Four of the blaCTX-Mgenes were sequenced. Statistical analysis carried out using SPSS (version 16) software and Chi-square test.Results:
High prevalent isolates were resistant to ampicillin, cephalothin, nalidixic acid and cotrimoxazole, but sensitive to imipenem, nitrofurantoin and amikacin. The ESBL phenotype was detected in 48 (51.6%) isolates, 42 of which were carrying blaCTX-Mgene. There was no significant difference between the β-lactamase-producing bacteria from the two studied cities. 81% and 85% of blaCTX-Mcarriers from hospitals in Qom and Mashhad were respectively found to have CTX-M-1 ESBL, whereas none of them showed CTX-M-2 ESBL cluster. BlaCTX-M-15was found in four selected bacteria by sequencing.Conclusions:
This study showed a high rate of CTX-M-1 cluster and also presence of CTX-M-15 ESBL in two different areas of Iran, which emphasize the need for employing an excellent management program in antibiotic therapy.Keywords
Beta-Lactamase CTX-M-15 Antibiotic Resistance Escherichia coli
1. Background
The accelerated increasing of antibiotic resistance is now emerging worldwide at an alarming rate among prevalent Gram negative bacteria, causing both community-acquired and nosocomial infections. One of the major causes of resistance in Enterobacteriaceae corresponds to resistance to broad-spectrum β-lactams, which is mainly associated with production of extended-spectrum β-lactamases (ESBLs) [1]. ESBLs are able to hydrolyze a broad spectrum of β-lactam antibiotics and their hydrolytic activity can be inhibited by several β-lactamase inhibitors such as clavulanic acid and tazobactam [1-3]. The first β-lactamase was identified in Escherichia coli prior to the release of penicillin for use in medical practice [4]. A variety of ESBLs, mostly of the CTX-M, TEM, and SHV types, have been reported in Enterobacteriaceae [5] and ESBLs currently display a major problem in antibiotic resistance in Enterobacteriaceae [6]. According to the references, CTX-M β-lactamases can divided into five groups based on their amino acid sequence identities. CTX-M-15 belongs to the CTX-M-1 cluster [7]. Phylogenetic studies suggest that blaCTX-Mgenes have evolved from chromosomal bla genes of Kluyvera sps [8, 9]. blaCTX-Mgene encodes a 291 amino acid enzyme although insertions and deletions in a few variants have sequences ranging from 282 to 292 amino acids. After the first 28 amino acids at the N-terminal are cleaved, the mature enzyme contains 263 amino acids [8]. blaCTX-Mis becoming more prevalent and over 125 different subtypes of the blaCTX-Mgene have been described with blaCTX-M-15most predominant around the globe associated with clonal spread [10].
Enterobacteriaceae are important opportunist pathogens and account for the majority of urinary tract infections and bacteremia [11]. E. coli is living in the intestinal tract of humans as mostly commensal reservoir, but some strains have the ability to create a wide variety of diseases in humans and some animals [12]. Urinary tract infection remains as one of the most prevalent infectious diseases worldwide leading to severe disorders like cystitis and pyelonephritis [13].
However there is a significant knowledge about antibiotic resistance but due to excessive use of antibiotics in Iran and ascending resistance in the communities, continuous research is necessary in this regard. Furthermore, there is a limited published data about the subtypes of ESBL genes among clinical isolates in some areas in Iran. This study was conducted to determine antibiotic susceptibility patterns and molecular properties of ESBL type CTX-M to detect blaCTX-M-15in E. coli urinary isolates from two different geographic regions in Iran by PCR and DNA sequencing.
2. Methods
2.1. Isolation and Identification of Bacteria
In this descriptive cross-sectional study, E. coli bacteria were isolated from non-duplicated urine specimens of hospitalized patients ages greater than 13 years, referred to two hospitals in two different geographic regions in Iran, Mashhad (Qaem and 17th Shahrivar hospitals) and Qom (Kamkar and Golpayegani), from August 2013 to February 2014. In this study, 102 clinical isolates of E. coli were isolated from middle urine. From these bacteria, 93 isolates were cultured in subsequent experiments in laboratory. Bacteria were identified phenotypically employing a combination of conventional identification methods, as well as the microgen identification system (Microgen Bioproducts GNA- ID UK).
2.2. Antibiotic Susceptibility Assay and Detection of ESBL Production
Antibiotic susceptibility of bacterial isolates to β-lactam and non-β-lactam antibiotics was carried out by disc diffusion method [14] using Mueller-Hinton agar (Merck, Germany) based on the Kirby-Bauer method and according to the criteria recommended by the clinical and laboratory standards institute (CLSI). Antibiotic agents were as follows: amikacin (AK, 30 µg), nalidixic acid (NA, 30 µg), ampicillin (AMP, 10 µg), cephalothin (KF, 30 µg), imipenem (IMI, 10 µg), co-trimoxazole (TS, 25 µg), cefepime (CPM, 30 µg), gentamicin (GM, 10 µg), nitrofurantoin (NIF, 300 µg), ceftazidime (CAZ, 30 µg), cefotaxime (CEP, 30 µg), cefpodoxime (CPD, 30 µg), cefixime (CFM, 5 µg), ceftriaxone (CRO, 30 µg). The discs were from MAST, UK (MAST DIAGNOSTICS-UK). E. coli ATCC 29522 was used for quality control. All tests were repeated three times.
ESBL production was detected using the double-disk synergy (DDS) test and ESBL confirmatory test [15] according to the CLSI criteria (CLSI, 2009). Double-disc synergy test (DDST) was carried out with amoxicillin-clavulanat (20 + 10 µg), cefotaxime (30 µg), ceftazidime (30 µg) [16]. ESBL production was defined when the inhibition zone of the disk of cephalosporin plus clavulanic acid was ≥ 5 mm greater than that of the disk of cephalosporin alone [17]. All experiments were repeated three times.
2.3. DNA Preparation and PCR Screening
Bacteria were grown in Luria Bertani broth to an OD600 of 1.0. Plasmid DNA of all isolates was extracted by miniprep kit (Perfect Prep-Spin Mini kit- 5 Prime- USA). Extracted plasmid DNA was targeted for the blaCTX-Mgene using the CTX-MU-1 and CTX-MU-2 primers described by Pagani, 2003 [18] which amplified a 593 bp fragment. PCR was performed under the following conditions: 94°C for 5 minutes, and 35 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds and a final extension in 72°C for 3 minutes in a thermal cycler (Kyratec- Korea). PCR products were visualized in agarose 1.5% w/v gels following electrophoresis and Green viewer staining. In this study E. coli strain producing ESBL type CTX-M prepared from Pasteur institute of Iran was used as positive control for blaCTX-Mgene.
CTX-M-1 and CTX-M-2 β-lactamase-producing genes were amplified using the primers and temperatures listed in Table 1. Reaction mixtures (30 µL) contained 3 µL (10X) of PCR buffer, 1 µL MgCl2 (10 mM), 0.25 µL Taq DNA polymerase (5 U/µL) (Fermentas-Lituania), 0.5 µL dNTPs (10 mM), 1 µL of each primer (10 µM) and 50 - 80 ng of each plasmid extracts. Five microliters of each PCR product was analyzed by electrophoresis in 1.5% (w/v) agarose along with a 100 bp ladder standard (Fermentas, Lithuania), positive and negative controls.
The Sequence of Primers and Temperatures Used in PCR blaCTX-M-1and blaCTX-M-2Genes
| Primers | Sequences (5’ → 3’) | Gen and Size | PCR | Cycles | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| CTX-M1GF | CGC TTT GCG ATG TGC AG’ | blaCTX-M1- 550 bp | 94°C- 30 s | 56°C- 30 s | 72°C- 30 s | 35 |
| CTX-M1GR | ACC GCG ATA TCG TTG GT | |||||
| CTX-M2GF | TTA ATG ACT CAG AGC ATT C’ | blaCTX-M2- 902 bp | 94°C- 30 s | 56°C- 45 s | 72°C- 45 s | 35 |
| CTX-M2GR | GAT ACC TCG CTC CAT TTA TTG | |||||
2.4. DNA Sequencing
Amplified DNAs of four samples were sequenced in both directions using CTX-MU-1 and CTX-MU-2 primers. Sequences were determined with an ABI 3730 DNA sequence by Lifesciences- Bioscience Company in UK. Sequence data were analyzed using the Sequencher sequence software alignment (Version 4.10.1) and were compared to the identified β-lactamase in the GenBank nucleotide database available on the Internet at the National center of biotechnology information website (http://www.ncbi.nlm.nih.gov).
2.5. Statistical Analysis
Statistical analysis carried out using SPSS (version 16) software. Chi-square test used for determination of significance association between two groups of bacteria; ESBL-producers and ESBL-no producers. Percentage of antibiotic resistance or sensitivity was determined according to the mean of inhibition zone and the P value ≤ 0.05 was considered significant.
3. Results
3.1. Bacteria
In this study, from a total of 93 E. coli bacteria isolated from patients in Mashhad and Qom hospitals, 49 isolates were from samples of patients in Qom and 44 isolates were from Mashhad.
3.2. Antibiotic Susceptibility Pattern and ESBL Producing Bacteria
All bacteria were susceptible to imipenem. Figures 1 and 2 show the antibiotic susceptibility pattern of E. coli isolates from hospitals in Mashhad and Qom, respectively. Resistant of isolated bacteria from urinary samples of patients in Mashhad was higher than of urinary isolates in Qom to the β-lactam antibiotics of ceftazidime, cephalothin, cefotaxime, cefepime, cefpodoxime and cefixime. The difference was significant for cephalothin (70.5% versus 49%). Significant difference was not observed between susceptibility of isolates from two areas to the non-β-lactam antibiotics (P > 0.05).
Based on the DDS test, 24 isolates were positive and 6 isolates were suspicious from 49 bacteria isolated in Qom. Among isolates from Mashhad, 21 were ESBL+ and 5 were suspicious. By ESBL confirmatory test, 48 of 93 isolated bacteria (51.6%) were ESBL-producing that 25 and 23 isolates of these bacteria were from Qom and Mashhad isolates, respectively. Thus, there was no significant difference between the β-lactamase-producing bacteria from the two studied cities (51% versus 53.5%).
Antibiotic Susceptibility Pattern of Urinary E. coli Isolates from Mashhad (AMP; Ampicillin, CAZ; ceftazidime, KF; Cephalothin, CEP; Cefotaxime, CPM; Cefepime, CRO; Ceftriaxone, CPD; Cefpodoxime, CFM; Cefixime, IMP; Imipenem, NA; Nalidixic Acid, AK; Amikacin, NIF; Nitrofurantoin, GM; Gentamicin, TS; Cotrimoxazole)
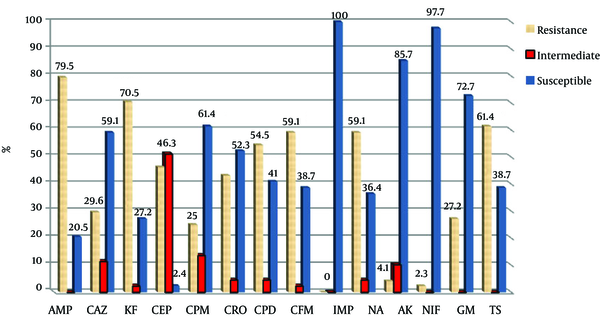
All β-lactamase-producing bacteria were identified using Microgen kit and were confirmed E. coli.
Antibiotic Susceptibility Pattern of Urinary E. coli Isolates from Qom (AMP; Ampicillin, CAZ; Ceftazidime, KF; Cephalothin, CEP; Cefotaxime, CPM; Cefepime, CRO; Ceftriaxone, CPD; Cefpodoxime, CFM; Cefixime, IMP; Imipenem, NA; Nalidixic Acid, AK; Amikacin, NIF; Nitrofurantoin, GM; Gentamicin, TS; Cotrimoxazole)
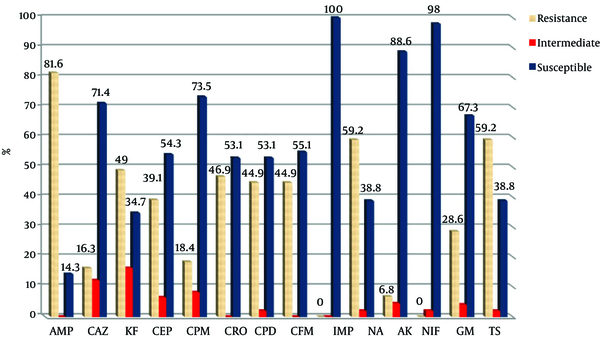
3.3. Results of Genetic Analysis
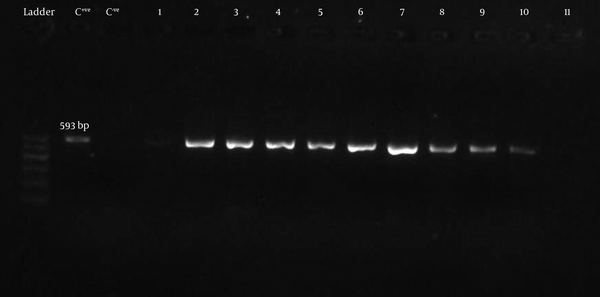
BlaCTX-Mgene was detected by PCR using universal primers (CTX-MU1 and CTX-MU2). Figure 3 shows the gel electrophoresis of PCR products for some bacteria. Twenty one of 23 (91.3%) ESBL producers from Mashhad and 21 of the 25 (84%) producers from Qom was carrying blaCTX-Mgene.
PCR Products of CTX-M Type ESBL Producing Isolates on 1.5% gel Agarose (C + ve; Positive Control, C-ve; Negative Control, Ladder; Marker 100 bp
Seventeen strains of 21 blaCTX-M carriers (81%) from hospitals in Qom, and 17 of 23 isolates (85%) from Mashhad was carrying blaCTX-M-1gene. However blaCTX-M-2gene was detected in none of the isolates.
Sequencing of the PCR products for two tested isolates from each city revealed that the sequences of isolates were similar to the compared strain in the GenBank (E. coli strain UE90CTXM blaCTX-M-15gene, with accession no. FJ997868). Figure 4 shows the result of sequencing of one isolate from Qom. The sequences reported therein have been deposited with GenBank database under accession numbers KJ739793, KJ739794, KJ739795 and KJ739796.
Sequencing of blaCTX-M-1PCR Product of One Isolate from Qom
4. Discussion
Our study revealed the highest sensitivity of isolates to imipenem, nitrofurantoin, and amikacin in both areas. The highest resistance was to ampicillin, cephalothin, and cotrimoxazole among isolates from Mashhad, while the highest resistance for isolates in Qom was to ampicillin, cotrimoxazole and nalidixic acid. The resistance to cephalothin in Mashhad was significantly higher than in Qom. Antibiotic resistance can vary depending on the geographic area or prevalence of nosocomial infections in different regions. A high percentage (51.6%) of the bacterial isolates was ESBL producing which can be a warning to health authorities in Iran. About 45.2% of isolates were carrying CTX-M-1 class that indicates the increasing prevalence of this type. The prevalence of CTX-M-1 had little difference among isolates from the two studied cities. BlaCTX-M-2gene was detected in none of the isolates.
E. coli as a common cause of urinary tract infections and bacteremia in humans is frequently resistant to aminopenicillins, such as amoxicillin or ampicillin, and narrow-spectrum cephalosporins. Resistance is typically mediated by the acquisition of plasmid-encoded β-lactamases, such as TEM, SHV and CTX-M-type enzymes [19].
Recent reports have shown a rapid and alarming dissemination of Enterobacteriaceae producing ESBLs of the CTX-M type in certain countries such as India [20], Cambodia [21], France [22], UK [23], Qatar [9] and Canada [24] that has become the most prevalent ESBL-type world-wide. A high prevalence and broad geographic distribution of CTX-M-producing E. coli and K. pneumonia nosocomial strains have been shown in Russia. The blaCTX-M type is the most common ESBL resistance gene in E. coli isolates in China, as most reports indicate [25]. β-lactamases of the CTX-M-1 cluster were the predominant CTX-M enzymes in both species. Out of this group only CTX-M-3- and CTX-M-15-like enzymes were identified among selected isolates [26]. However, we found a higher prevalence of CTX-Ms in selected area in Iran than that reported in many of these studies.
44.5% (89 of 200) of E. coli isolates from five hospitals in Tehran carried the blaCTX-M-1group alleles in 2009 to 2010. Imipenem and amikacin were effective against 100% and 85% of all tested isolates respectively [27]. The result of this research was very similar to our study results. A study in 2012 showed a high rate of CTX-M-1 cluster - ESBLs in South-Eastern Nigeria and further confirms the worldwide spread of CTX-M ESBLs in clinical isolates and among them, in three exemplary strains the CTX-M-15 ESBL was confirmed [28]. BlaCTX-M-15, which belongs to the CTX-M-1 cluster, has been reported worldwide. A study in a French hospital between 2006 and December 2010 has reported an increase in the prevalence of ESBL-producing strains among E. coli isolates recovered from neonates or their mothers in which rose from 1.15% (6/523) in 2006 to 4.1% (22/543) in 2010. BlaCTX-M15was one of the principal ESBLs found in this study with prevalence 18.8% [29].
E. coli isolates carried the CTX-M-1 type ESBLs. In our study CTX-M-15 ESBL, was confirmed in four selected isolates. This result can emphasize that this enzyme is now one of the most common CTX-M β-lactamases in Iran. In several countries CTX-M-15 is currently the most frequent ESBL in E. coli like in the United Kingdom (UK), India, Spain, France, Latin America and Lebanon [28]. The gene of blaCTX-M-15has been reported in Klebsiella pneumonia isolated from hospitals of Tehran in 2014 [30]. Among 1168 ESBL- E. coli isolates obtained from various clinical specimens in China, 96.2% of the isolates were detected to harbor blaCTX-Mgenes. BlaCTX-M-1group and blaCTX-M-9group were detected at 40.7% and 48.7% respectively. BlaCTX-M-55(24.8%) and blaCTX-M-15(18.2%) were the major genotypes in blaCTX-M-1group while blaCTX-M-14(46.8%) as predominant in blaCTX-M-9group [31]. Also, E. coli CTX-M-15 has been reported from bovine mastitis in the UK that emphasizes the clinical importance of detecting ESBL-producing bacteria in animal food products and the need for routine screening in veterinary samples [32].
Our findings suggest that CTX-M-type β-lactamases are widespread in selected areas in Iran. Sequencing of the PCR products for two examined isolates from each city revealed that the sequence of isolates was similar the CTX-M-15 type β-lactamase. The results emphasize the need for employing a regular monitoring of antibiotic resistance and also an excellent management program in antibiotic therapy in our country.
Acknowledgements
References
-
1.
Nordmann P, Dortet L, Poirel L. Rapid detection of extended-spectrum- -lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2012;50(9):3016-22. https://doi.org/10.1128/jcm.00859-12.
-
2.
Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969-76. [PubMed ID: 19995920]. https://doi.org/10.1128/AAC.01009-09.
-
3.
Markovska R, Schneider I, Keuleyan E, Sredkova M, Ivanova D, Markova B, et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in Bulgarian hospitals. Microb Drug Resist. 2008;14(2):119-28. [PubMed ID: 18489242]. https://doi.org/10.1089/mdr.2008.0814.
-
4.
Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146(3713):837.
-
5.
Poirel L, Bonnin RA, Nordmann P. Genetic support and diversity of acquired extended-spectrum beta-lactamases in Gram-negative rods. Infect Genet Evol. 2012;12(5):883-93. [PubMed ID: 22414916]. https://doi.org/10.1016/j.meegid.2012.02.008.
-
6.
Rodriguez-Bano J, Pascual A. Clinical significance of extended-spectrum beta-lactamases. Expert Rev Anti Infect Ther. 2008;6(5):671-83. [PubMed ID: 18847405]. https://doi.org/10.1586/14787210.6.5.671.
-
7.
Owlia P, Bameri Z, Chitsaz M. Detection of CTX-M-β lactamases form isolated klebsiella pneumoniae. Iranian Journal of Pathology. 2010;5(3):137-42.
-
8.
Sridhar Rao PN. Extended spectrum beta-lactamases- A comprehensive review. 2015. Available from: www.microrao.com/micronotes/pg/esbl_review.pdf.
-
9.
Sid Ahmed MA, Bansal D, Acharya A, Elmi AA, Hamid JM, Sid Ahmed AM, et al. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob Resist Infect Control. 2016;5:4. [PubMed ID: 26865975]. https://doi.org/10.1186/s13756-016-0103-x.
-
10.
Sullivan R, Schaus D, John M, Delport JA. Extended spectrum beta-lactamases: A minireview of clinical relevant groups. J Med Microb Diagn. 2015;4(203):2161-703.1000203.
-
11.
Livermore DM, Woodford N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 2006;14(9):413-20. [PubMed ID: 16876996]. https://doi.org/10.1016/j.tim.2006.07.008.
-
12.
Tille P. Bailey & Scott's diagnostic microbiology. Elsevier Health Sciences; 2015.
-
13.
Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, et al. Virulence characteristics and genetic affinities of multiple drug resistant uropathogenic Escherichia coli from a semi urban locality in India. PLoS One. 2011;6(3). e18063. [PubMed ID: 21464963]. https://doi.org/10.1371/journal.pone.0018063.
-
14.
Cockerill FR. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. Clinical and Laboratory Standards Institute (CLSI); 2011.
-
15.
Saeidi S, Alavi-Naini R, Shayan S. Antimicrobial susceptibility and distribution of tem and ctx-m genes among ESBL-producing Klebsiella pneumoniae and Pseudomonas aeruginosa causing urinary tract infections. Zahedan J Res Med Sci. 2014;16(4):1-5.
-
16.
M'Zali FH, Chanawong A, Kerr KG, Birkenhead D, Hawkey PM. Detection of extended-spectrum beta-lactamases in members of the family enterobacteriaceae: comparison of the MAST DD test, the double disc and the Etest ESBL. J Antimicrob Chemother. 2000;45(6):881-5. [PubMed ID: 10837444].
-
17.
Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46(1):1-11. [PubMed ID: 11751104].
-
18.
Pagani L, Dell'Amico E, Migliavacca R, D'Andrea MM, Giacobone E, Amicosante G, et al. Multiple CTX-M-type extended-spectrum beta-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol. 2003;41(9):4264-9. [PubMed ID: 12958255].
-
19.
Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6 Suppl 1):S3-10. discussion S62-70. [PubMed ID: 16735149]. https://doi.org/10.1016/j.amjmed.2006.03.011.
-
20.
Canton R, Coque TM. The CTX-M beta-lactamase pandemic. Curr Opin Microbiol. 2006;9(5):466-75. [PubMed ID: 16942899]. https://doi.org/10.1016/j.mib.2006.08.011.
-
21.
Ruppe E, Hem S, Lath S, Gautier V, Ariey F, Sarthou JL, et al. CTX-M beta-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis. 2009;15(5):741-8. [PubMed ID: 19402960]. https://doi.org/10.3201/eid1505.071299.
-
22.
Arpin C, Dubois V, Coulange L, Andre C, Fischer I, Noury P, et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in community and private health care centers. Antimicrob Agents Chemother. 2003;47(11):3506-14. [PubMed ID: 14576109].
-
23.
Woodford N, Ward ME, Kaufmann ME, Turton J, Fagan EJ, James D, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother. 2004;54(4):735-43. [PubMed ID: 15347638]. https://doi.org/10.1093/jac/dkh424.
-
24.
Pitout JD, Hanson ND, Church DL, Laupland KB. Population-based laboratory surveillance for Escherichia coli-producing extended-spectrum beta-lactamases: importance of community isolates with blaCTX-M genes. Clin Infect Dis. 2004;38(12):1736-41. [PubMed ID: 15227620]. https://doi.org/10.1086/421094.
-
25.
Shi H, Sun F, Chen J, Ou Q, Feng W, Yong X, et al. Epidemiology of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing nosocomial -Escherichia coli infection in China. Ann Clin Microbiol Antimicrob. 2015;14:4. [PubMed ID: 25591816]. https://doi.org/10.1186/s12941-015-0063-7.
-
26.
Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47(12):3724-32. [PubMed ID: 14638473].
-
27.
Peerayeh SN, Eslami M, Memariani M, Siadat SD. High prevalence of blaCTX-M-1 group extended-spectrum β-lactamase genes in Escherichia coli isolates from Tehran. Jundishapur J Microbiol. 2013;6(7).
-
28.
Iroha IR, Esimone CO, Neumann S, Marlinghaus L, Korte M, Szabados F, et al. First description of Escherichia coli producing CTX-M-15- extended spectrum beta lactamase (ESBL) in out-patients from south eastern Nigeria. Ann Clin Microbiol Antimicrob. 2012;11:19. [PubMed ID: 22824236]. https://doi.org/10.1186/1476-0711-11-19.
-
29.
Birgy A, Mariani-Kurkdjian P, Bidet P, Doit C, Genel N, Courroux C, et al. Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: prevalence of E. coli ST131. J Clin Microbiol. 2013;51(6):1727-32.
-
30.
Hashemi A. Detection of blactx-m, blatem, blashv genes in Klebsiella pneumoniae strains isolated from two hospitals of Tehran, Iran. Iran J Public Health. 2014;43(2):98.
-
31.
Xia S, Fan X, Huang Z, Xia L, Xiao M, Chen R, et al. Dominance of CTX-M-type extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS One. 2014;9(7). e100707. [PubMed ID: 24983621]. https://doi.org/10.1371/journal.pone.0100707.
-
32.
Timofte D, Maciuca IE, Evans NJ, Williams H, Wattret A, Fick JC, et al. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 beta-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob Agents Chemother. 2014;58(2):789-94. [PubMed ID: 24247146]. https://doi.org/10.1128/AAC.00752-13.