Abstract
Background:
Because survivin has a significant role as an inhibitor in breast cancer (BC) advancement and development and its productivity and interactions may well be under the effects of polymorphisms in the gene promoter we investigated a common polymorphism (-31G/C) that has been reported to be influential on the survivin expression in different forms of cancer. But the observations did not show any correlation between this SNP (single nucleotide polymorphism) and breast cancer disease at least in our study population (east Azerbaijan, Iran). The (-31G/C polymorphism is the cause of over expression of survivin once it de-represses the cell cycle-dependent transcription of survivin gene.Objectives:
In this paper, the aim has been to study the role of -31G/C in breast cancer in the hospitals of the province of east Azerbaijan, Iran.Patients and Methods:
In this case-control study, 5 mL of peripheral blood samples of 82 healthy subjects and 94 breast cancer patients with pathologically confirmed were gathered. We analyzed -31G/C single nucleotide polymorphisms of survivin promoter with using of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and gene sequencing. The required samples were acquired from two of the general hospitals in Tabriz.Results:
Statistical analysis revealed that genotype frequencies and allele distribution of the survivin promoter -31G/C for both controls and cases were similar. There has been no statistically noteworthy association between breast cancer and the variant genotypes (CC and CG).Conclusions:
Seemingly, there has been no statistically association between -31G/C polymorphism with BC and clinicopathological characteristics in the population being investigated in this paper.Keywords
Association Polymorphism Breast Cancer Promoter Survivin Gene
1. Background
Breast cancer (BC) has been reported to be the most common fatality and the second cause of death by cancer among women in the world. Thanks to the timely detection and effective treatment, the death rate is on the decline. Nonetheless, metastasis still poses as an alarming factor leading to the failure of therapies and patients’ death [1].
Apoptosis is a key factor in homeostasis of organs and cells. Thus, defects in the regulation of apoptosis can lead to many human disorders such as cancers. Programmed cell death or apoptosis is carried out through a cascade of enzymes known as caspases. IAPs (inhibitors of apoptosis proteins) that prohibit caspases, including initiator and effector caspases (caspase 9, caspase 2 and caspase 3) are a family of anti-apoptotic proteins. So far, eight human IAP family members with one to three baculovirus IAP repeat domains and a C-terminal RING or CARD (Caspase activation recruitment domain) domain have been recognized [2]. Survivin or BIRC5 (baculoviral inhibitor of apoptosis repeat-containing 5) is a member of IAP family which in is encoded as the BIRC5 gene in humans. Survivin is possibly the most unique member if IAP in terms of structure and has only a single BIR domain which is crucial because of its anti-apoptotic behavior. Proteins being encoded by this gene lack a c-terminus RING finger or CARD domain [3, 4]. The gene coding for survivin is fundamental on human chromosome 17q25, and consists 142 amino acids [3]. It has a canonical CpG island in the promoter region and numerous Sp1 sites in the 5’flanking region and lacking a TATA box motif [5].
Survivin gene is usually expressed in embryonic tissues and is up regulated in many human cancers including breast cancer, thus playing a critical role in carcinogenesis [3]. Null mutations of survivin leads to early embryonic death, showing its key role in cell development, differentiation and homeostasis [5]. It is therefore crucial for cancer cell survival due to its significant role in regulation of both apoptosis and mitosis processes [1, 5]. As a result, survivin becomes a target for cancer therapy for its influence in the process mentioned above. northwest of Iran is comparably a highly potential region for BC incidences. Recent investigations indicate that promoter elements including CDE/CHR regulate the expression of survivin during G2/M phase of mitosis. The -31G/C SNP is placed inside the CDE/CHR repressor binding site. Provided that this motif is functionally disrupted, cell cycle- dependent transcription of survivin will be altered [3, 5-8]. Alterations in gene expression are mainly due to genetic variations, especially the regulatory polymorphisms located in promoter regions. In cancer cells overexpression of survivin at both protein and mRNA levels is linked to genetic variant -31G/C in the survivin promoter. Lots of studies have been focused on this polymorphism and its role on risk of gastric urothelial, colorectal, cervical and lung cancers. However the role of genetic variants in the survivin promoter region on clinical results has been reported by few of researchers [5].
2. Objectives
This paper aims at investigation of the link between -31G/C SNP and BC in an Iranian population of east Azerbaijan.
3. Patients and Methods
In this case-control study, 176 individuals (94 breast cancer cases and 82 healthy controls, that all of them were women) were investigated and the samples were obtained from Imam Reza and Nour-Nejat hospitals in Tabriz (Iran) and informed consent was obtained. In a period of two years from 2008 to 2010 clinicopathological data of patients, without prior treatment, from medical records were obtained. The first and second level family members and relatives were check for the presence of the disease in the history of the family. Blood samples were obtained in EDTA (Ethylenediaminetetraacetic acid) from each individual and were preserved frozen until the extraction of DNA. Genomic DNA was extracted from peripheral blood samples using proteinase K digestion and phenol/chloroform method.
3.1. Polymorphism Genotyping
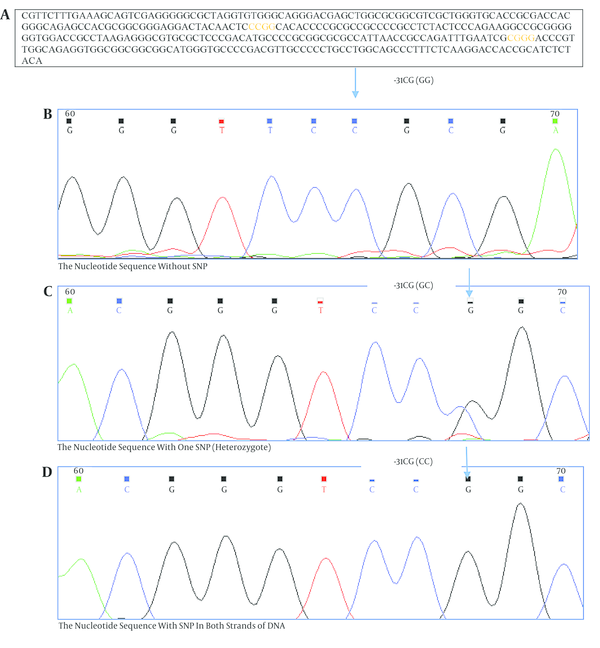
To identify genetic variants in the survivin gene promoter for screening of -31G/C polymorphism, a 329 bp region of promoter was sequenced. The survivin -31G/C polymorphism was then analyzed by polymerase chain reaction- restriction fragment containing the -31 polymorphic site was amplified using two primers; the forward primer 5’-CGTTCTTTGAAAGCAGTCGAG-3’ and the reverse primer: 5’-AGGACCACCGCATCTCTACA-3’ (Table 1) [9].
Conditions for Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) Analysis of the Survivin Promoter
| Position of SNP | Primers | Annealing Temperature | PCR Product, bp | Enzyme | Digested PCR Products |
|---|---|---|---|---|---|
| 58°C | 329 | MSPI | GG: 214, 115 | ||
| F | 5’CGTTCTTTGAAAGCAGTCGAG3’ | GC: 214, 120, 115, 94 | |||
| R | 5’TGTAGAGATGCGGTGGTCCT3’ | CC: 120, 115, 94 |
Each PCR reaction was performed in total reaction volume of 25 μL containing 2 μL 10x PCR buffer, 0.8 μL Mgcl2, 0.2 μL dNTP, 1 μL of each primer, 50 ng genomic DNA and 0.18 μL Tag DNA polymerase.
3.2. The Conditions of Polymerase Chain Reaction (PCR)
initial denaturation step at 95°C for 2 minutes, followed by 32 cycles of 95°C for 20 seconds as the second melting step, 58°C for10 seconds for primer annealing and extension at 72°C for 20 seconds and a final extension step of 72°C for 3 minutes. Ten microliters of the PCR products were digested with 5 UMSPI and 2 μL NEBuffer4 (New England biolabs) at 37°C over night. The digested products were separated on a 10% acrylamid gel for 2 hours and stained with silver nitrate. Eleven individuals were checked by the patterns of the digested PCR products and about 8% randomly were rechecked for quality control and the concordance was 100%.
The final collected data were scrutinized by SPSS-15 software through the Pearson χ2 and ANOVA tests. The link between -31G/C polymorphism of survivin and risk of breast cancer was predicted by odd ratios and 95% confidence intervals. The homozygous genotype GG in the control group for the common allele formed the reference group. The compared groups indicated a probability of 7% which is quite significant.
4. Results
Results of this study (Table 2) demonstrate the survivin genotype and allele frequency distribution among the 94 BC patients and 82 healthy controls are demos. The prevalence of C/G, G/G genotypes was significantly greater in two groups than C/C genotype.
Genotype and Allele Distribution of the -31 Survivin Promoter for Both Controls and Cases
| Groups | 95% CI (Genotype as a Grouping Variable) | P Value | |||
|---|---|---|---|---|---|
| Cases | Controls | P Value | |||
| 0.84 | 0.31 | ||||
| CC | 3 (3.2) | 0.42 (-2.48 - 3.30) | |||
| CG | 44 (46.8) | 0.28 (0.16 - 0.41) | |||
| GG | 47 (50) | 0.43 (0.28 - 0.57) | |||
| NA | 0.86 | ||||
| C | 45 [0.27(0.21 - 0.34)] | NA | |||
| G | 119 [0.72(0.65 - 0.78)] | NA | |||
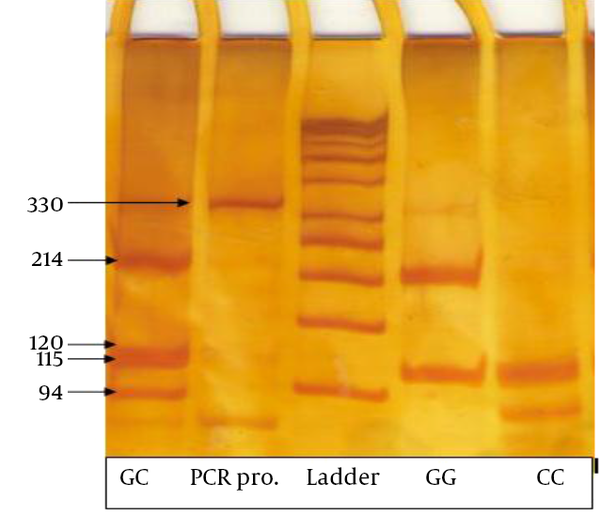
Genotypes were considered as follows: two fragments of 214 and 115 bp for the GG genotype, three fragments of 120, 115 and 94 bp for CC genotype, and four fragments of 214, 120, 115 and 94 bp for the CG genotype (Figures 1 and 2).
The Restriction Fragment Length Polymorphism (RFLP) Products on 10% Acrylamide Gel and Genotype Patterns From Three Cases
The Nucleotide Sequences of PCR Products of Cases
The results of analysis for association between risk of BC and the survivin -31G/C SNP are summarized in Tables 3 and 4.
Association of Selected Characteristics From BC Patients With BIRC5 -31G/C Polymorphism
| Stage (95% CI) | P Value | |||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 3.05 (-2.25 - 8.35) | 2.87 (1.29 - 4.45) | 4.3 (2.76 - 5.83) | 0.21 | |
| 42 (35.08 - 48.92) | 46.11 (40.32 - 51.89) | 48.21 (45.36 - 51.06) | 0.20 | |
| 0.98 | ||||
| Left | 4 (12.1) | 8 (24.2) | 21 (63.6) | |
| Right | 5 (11.9) | 11 (26.2) | 26 (61.9) | |
| 0.03 | ||||
Association of the Survivin Genotypes With the Tumor Stage and Side of the Breast Involvement
| Genotype Frequency | Side Frequency, % | P Value | 95% CI | |||
|---|---|---|---|---|---|---|
| CC | CG | GG | ||||
| 0.97 | ||||||
| Left | 1 (2.5) | 19 (47.5) | 20 (50) | 44.4 | Na | |
| Right | 2 (4) | 24 (48) | 24 (48) | 55.6 | Na | |
| 0.73 | ||||||
| 1 | 0 (0) | 6 (17.6) | 3 (7.9) | 12 | Na | |
| 2 | 2 (66.7) | 6 (17.6) | 11 (28.9) | 25.3 | 0.11 (0.05-0.17) | |
| 3 | 1 (33.3) | 22 (64.7) | 24 (63.2) | 62.7 | 0.55 (0.44-0.67) | |
No discrepancy related to age between patients and controls has been identified. The ages of individuals ranged from 21 to 72 years. Approximately 95% of the patients were older than 30 years (Table 5). Similarly, no association was observed stratified by the left or right side status of breast. Nonetheless, an association was found between tumor stage (2 and 3) and lymph nod metastasis (P < 0.01). Seventeen percent of patients did not reveal nodal involvement. The target population in this paper did not reveal a significant association between genotypes and clinicopathological features of BC.
| Genotype | Age, y | ||||
|---|---|---|---|---|---|
| < 30 | 31 - 40 | 40 - 50 | 50 - 60 | > 60 | |
| 2 (4.5) | 8 (18.2) | 16 (36.4) | 11 (25.0) | 7 (15.9) | |
| 0 (0) | 15 (35.7) | 15 (35.7) | 8 (19.0) | 4 (9.5) | |
| 0 (0) | 0 (0) | 2 (66.7) | 1 (33.3) | 0 (0) | |
Applying RFLP, we determined the nucleotide variants at the polymorphic site of survivin gene promoter. No differential distribution was observed between patients with BC and controls. Concerning the tumor features, no important association was found between -31G/C polymorphism and tumor size (Table 6), stage, side, pathology and age pattern as well. The frequency of -31G/C polymorphism within the survivin promoter was not significantly different between BC samples and healthy individuals.
Association of Genotypes With Tumor Size
| Tumor Size, cm | Genotype Frequency | Size Frequency | P Value | ||
|---|---|---|---|---|---|
| CC | CG | GG | |||
| 0 | 53.3 | 66.7 | 57.1 | 0.79 | |
| 100 | 33.3 | 16.7 | 28.6 | 0.79 | |
| 0 | 13.3 | 16.7 | 14.3 | 0.79 | |
5. Discussion
In the present study, the association between survivin -31 genotypes and breast cancer was investigated. No discrepancy in distribution of these genotypes between the studied populations was identified. However, we observed an association between the stage 2 and 3 of tumor with nodal involvement in our study population. Similar studies by Xu et al. [10] indicated that the -31 polymorphism in the promoter of survivin was not involved in the process of development of cervical cancer in Hungary. Controversially, Jang et al. [2] reported an association between this polymorphism and lung cancer risk [11]. Also Borbely et al. [12] demonstrated that -31G/C SNP up-regularized the survivin expression at protein levels as well as mRNA levels [11]. Wagner et al. reported that -31G/C is a functional polymorphism in survivin promoter region [9]. Our result somehow is in line with the result of Yang et al. study [6] which showed that the survivin -31G/C genotype distribution in esophageal squamous cell cancer patients and healthy controls was not statistically significant. In another research the risk of esophageal squamous cell carcinoma, has been reported to be associated with polymorphisms of survivin promoter [2].
There are five common polymorphisms in the survivin promoter region, in which -31G/C (rs9904341) is most widely studied, because its location is at the CDE/CHR repressor elements so it could be functional. It has been demonstrated by researchers that this SNP is linked with increased expression of survivin gene and aberrant cell cycle-dependent transcription [5]. There are no reports on similar studies on breast cancer; however, our finding on this SNP was considered preliminary and further larger studies are needed to validate the significance of this SNP. Survivin is a valuable marker in the prognosis and genetic of cancer because of its differential expression in many malignancies [3]. Also it is the smallest member of the IAP protein family which regulates cell division and can be initiated via two pathways, the extrinsic or intrinsic apoptotic pathways [7, 8, 13]. Survivin or BIRC5 gene is expressed either in embryonic and fetal organs or in the majority of human cancers (breast, lung, colon, pancreas, bladder, stomach, ovarian, oral and gastric cancer), but usually is not detectable in normal adult tissues with the exception of thymus, basal colonic epithelial and endothelial cells [9, 14].
Survivin is greatly controlled at the transcription level and is expressed in G2/M phase of the cell cycle. CDE (GGCGG) and CHR (ATTTGAA) repressor elements which are located in the proximal region of the survivin promoter mediate its transcription. This gene has a lot of roles such as chromosomal attachment, S-phase progression, inhibition of caspases and etc. [7, 8]. Polymorphism of -31G/C which located at the CDE/CHR repressor elements in the promoter region likely de-repress survivin transcription in the G1 phase in various cancers. Many studies have shown that this SNP may be a functional SNP within the promoter [5, 13, 14]. By functional disruption of CDE/CHR motifs at the binding site, the occurrence of this SNP was correlated with the over expressing of survivin at both the mRNA and protein levels [5, 7, 8]. For finding a useful marker to identify genetic susceptibility, polymorphism of surviving gene might be of help since survivin acts as an inhibitor of apoptosis which is essential for eliminating mutated or transformed cells from the body, thus it may be possible that subjects with a lower production genotype for survivin have increased apoptotic capacity to eliminate cells with DNA damage and have decreased susceptibility to breast cancer. This polymorphism may depress survivin transcription by modifying the binding motif of the CDE/CHR repressor [2].
Contrary to our expectations in this study, we could not find any substantial discrepancy in the genotype distributions of the -31G/C variants among two compared groups. Thereby, because SNPs often is different between ethnic populations, further studies with larger sample size are needed to verify the association of survivin promoter polymorphism with BC. Our results showed that the frequency of the C/C and G/G genotypes was 3.2% and 50% in patients with BC and 4.9% and 50% in controls, respectively.
We can conclude that the survivin -31G/C SNPs were not correlated to BC in our study population. It seems that to clarify the mechanisms of promoter region of survivin in BC we need a large sample with diverse ethnic groups to verify the role of functional SNPs in the development of BC and their associations with clinicopathological factors.
We discovered that the -31C > G polymorphism in the survivin gene was not correlated to the genetic susceptibility to breast cancer. Although more studies with larger sample size are needed to confirm our findings. In addition, cause genetic polymorphisms often are different between ethnic groups, further studies should be done to clarify the association between survivin polymorphisms and Breast cancer in diverse ethnic groups.
References
-
1.
Peng XC, Yang L, Yang LP, Mao YQ, Yang HS, Liu JY, et al. Efficient inhibition of murine breast cancer growth and metastasis by gene transferred mouse survivin Thr34-->Ala mutant. J Exp Clin Cancer Res. 2008;27:46. [PubMed ID: 18816410]. https://doi.org/10.1186/1756-9966-27-46.
-
2.
Jang JS, Kim KM, Kang KH, Choi JE, Lee WK, Kim CH, et al. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60(1):31-9. [PubMed ID: 17961802]. https://doi.org/10.1016/j.lungcan.2007.09.008.
-
3.
Upadhyay R, Khurana R, Kumar S, Ghoshal UC, Mittal B. Role of survivin gene promoter polymorphism (-31G>C) in susceptibility and survival of esophageal cancer in northern India. Ann Surg Oncol. 2011;18(3):880-7. [PubMed ID: 20957442]. https://doi.org/10.1245/s10434-010-1371-y.
-
4.
Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, et al. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58(23):5315-20. [PubMed ID: 9850056].
-
5.
Han CH, Wei Q, Lu KK, Liu Z, Mills GB, Wang LE. Polymorphisms in the survivin promoter are associated with age of onset of ovarian cancer. Int J Clin Exp Med. 2009;2(4):289-99. [PubMed ID: 20057973].
-
6.
Yang X, Xiong G, Chen X, Xu X, Wang K, Fu Y, et al. Polymorphisms of survivin promoter are associated with risk of esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135(10):1341-9. [PubMed ID: 19352701]. https://doi.org/10.1007/s00432-009-0575-7.
-
7.
Yang L, Zhu H, Zhou B, Gu H, Yan H, Tang N, et al. The association between the survivin C-31G polymorphism and gastric cancer risk in a Chinese population. Dig Dis Sci. 2009;54(5):1021-8. [PubMed ID: 18716870]. https://doi.org/10.1007/s10620-008-0441-5.
-
8.
Cheng ZJ, Hu LH, Huang SJ. [Correlation of -31G/C polymorphisms of survivin promoter to tumorigenesis of gastric carcinoma]. Ai Zheng. 2008;27(3):258-63. [PubMed ID: 18334114].
-
9.
Wagner M, Schmelz K, Dorken B, Tamm I. Epigenetic and genetic analysis of the survivin promoter in acute myeloid leukemia. Leuk Res. 2008;32(7):1054-60. [PubMed ID: 18206228]. https://doi.org/10.1016/j.leukres.2007.11.013.
-
10.
Xu Y, Fang F, Ludewig G, Jones G, Jones D. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004;23(7):419-29. [PubMed ID: 15294091]. https://doi.org/10.1089/1044549041474788.
-
11.
Gazouli M, Tzanakis N, Rallis G, Theodoropoulos G, Papaconstantinou I, Kostakis A, et al. Survivin -31G/C promoter polymorphism and sporadic colorectal cancer. Int J Colorectal Dis. 2009;24(2):145-50. [PubMed ID: 18946675]. https://doi.org/10.1007/s00384-008-0601-2.
-
12.
Borbely AA, Murvai M, Szarka K, Konya J, Gergely L, Hernadi Z, et al. Survivin promoter polymorphism and cervical carcinogenesis. J Clin Pathol. 2007;60(3):303-6. [PubMed ID: 16714396]. https://doi.org/10.1136/jcp.2006.037804.
-
13.
Yang X, Xiong G, Chen X, Xu X, Wang K, Fu Y, et al. Survivin expression in esophageal cancer: correlation with p53 mutations and promoter polymorphism. Dis Esophagus. 2009;22(3):223-30. [PubMed ID: 19018856]. https://doi.org/10.1111/j.1442-2050.2008.00885.x.
-
14.
Wang YH, Chiou HY, Lin CT, Hsieh HY, Wu CC, Hsu CD, et al. Association between survivin gene promoter -31 C/G polymorphism and urothelial carcinoma risk in Taiwanese population. Urology. 2009;73(3):670-4. [PubMed ID: 19038421]. https://doi.org/10.1016/j.urology.2008.09.048.