Abstract
Background:
Detection of anti-HLA antibodies (HLAabs) in organ transplantation recipients is vital to determine whether the recipient has specific HLAabs against the donor’s HLA molecules (donor-specific anti-HLA antibodies [DSA]).Methods:
This preliminary study involved seven subjects: five without prior HLA sensitization and two with a history of possible HLA sensitization (kidney transplantation and pregnancy). Two of the subjects were siblings, two of the subjects were mother and son, and two of them were first cousins once removed. All the subjects underwent HLA typing, and four of the subjects underwent HLAabs assays using the sequence-specific oligonucleotide probes (SSOP) method.Results:
There were 10/16 HLA matches between subjects who were siblings and between subjects who were mother and son, and 6/16 HLA matches were found between subjects who were first cousins once removed. Subjects with previous kidney transplantation developed 59 HLA-Abs toward class I and class II HLA (the mean fluorescence intensity (MFI) value range: 1,003.36 to 12,404.77). HLAabs were also found among subjects without prior HLA sensitization, albeit with relatively lower MFI values.Conclusions:
HLA typing and HLAabs detection for donor-recipient compatibility assessment using the SSOP method offers more detailed, semi-quantitative results. Further research involving larger subjects in cohort settings will be useful for profiling MFI cut-off values for HLAabs.Keywords
Kidney Transplantation Chronic Kidney Disease (CKD) Histocompatibility Testing HLA
1. Background
Despite all the benefits of kidney transplantation (KT) as a treatment for end-stage renal disease (ESRD), this procedure also has its downsides. KT is not devoid of complications, such as infections, blocked ureter, urine leakage, and rejection. Rejection is defined as the immune system response to attack the allograft kidney tissue because it is recognized as a foreign tissue.
The gene that mediates graft rejection is HLA. The HLA molecules are unique for each individual and serve as markers, which help to tell the difference between self and non-self molecules. There are three classical loci at HLA class I: HLA-A, -B, and -C, and five loci at class II: HLA-DR, -DQ, -DP, -DM, and -DO. The system is highly polymorphic (1). Although class I antigens (HLA-A, -B, and -C) are the primary targets of alloantibodies, emerging evidence indicates that antibody reactivity to class II antigens, especially HLA-DR and HLA-DQ, may also result in graft loss and rejection (2).
Concerning donor-recipient compatibility, more HLA molecules shared between a donor and a recipient make it less likely for rejection to occur. Therefore, it is critical for a recipient candidate to find a donor whose HLA molecules are as compatible as possible to those of the recipient. To achieve this purpose, it is vital to examine HLA molecules of a donor and a recipient by performing HLA typing before transplantation (1, 2). In addition to performing HLA typing, the identification of HLA-Abs is also significant in organ transplantation because HLA typing alone cannot determine if the recipient has already formed HLA-specific antibodies to certain antigens. This could be acquired through transfusion, pregnancy, or previous organ transplantation, all of which are common among patients with ESRD, and will be re-exposed by implantation of the graft kidney (3).
2. Objectives
This study aimed to conduct an initial experiment to perform HLA typing and HLA-Abs examination to assess donor-recipient compatibility for KT in Indonesia.
3. Methods
3.1. Patient Selection
We included seven subjects in this pilot study: five subjects without any history of HLA sensitization and two subjects with suspected prior HLA sensitization. Five subjects were recruited voluntarily for this study and were not candidates for KT, while two subjects (subjects 6 and 7) were a donor-recipient candidate pair for KT (Table 1).
| Subject | Age, y | Gender | Relationship | History of HLA Exposure |
|---|---|---|---|---|
| 1 | 43 | Male | Siblings | None |
| 2 | 23 | Female | None | |
| 3 | 44 | Male | None | None |
| 4 | 25 | Male | Son and mother | None |
| 5 | 60 | Female | Pregnancy | |
| 6 | 65 | Male | First cousins once removed | Previous kidney transplantation |
| 7 | 25 | Male | None |
Subject 6 was diagnosed with ESRD six years before recruitment in the study. The subject received hemodialysis treatment for seven months before undergoing first KT surgery. Six years after the first KT, Subject 6 was diagnosed with graft failure and became a candidate for a second KT surgery. Subject 6 never had blood transfusion during the course of his illness.
All the subjects were recruited from the Urology Clinic at Dr. Cipto Mangunkusumo Hospital (Jakarta, Indonesia) from June-December 2016. We explained the study procedure to the subjects, and they signed the informed consent form. The Ethical Committee of the Faculty of Medicine, Universitas Indonesia, approved the study.
3.2. Study Design
We conducted HLA typing on all the subjects and HLA-Abs examination on four of the subjects (subjects 1, 3, 4, and 6). We performed both HLA typing and HLA-Abs examination using the SSOP method on a Luminex® platform. The same assessors unaware of the relationship between the subjects and whether the subjects had previous exposure to HLA molecules, carried out all examinations at Laboratorium Terpadu-Diagnostic and Research Center (DIARC) at the Faculty of Medicine Universitas Indonesia (Jakarta, Indonesia).
We compared the result of the HLA typing examination on both class I (HLA-A, -B, and -C) and class II (HLA-DR, -DP, and -DQ) HLA molecules between all the subjects, emphasizing the number of HLA matches/mismatches between the subjects with familial relationship. The result of the HLA typing examination was presented as the HLA class, locus, and the most detailed allele detected with the Luminex® machine. Furthermore, we compared the HLA-Abs examination result between the subjects who were examined, particularly between Subjects 6 and 7, for possible DSA finding. Apart from HLA typing, we also performed an HLA antibody examination in each subject. HLA antibodies towards their respective HLA antigens in each subject were presented according to each antibody's mean fluorescence intensity (MFI) from the highest to the lowest value.
4. Results
The age range of the subjects was 23-65 years. Subjects 1 and 2 (siblings) had 10/16 matches of HLA molecules. Similarly, subjects 4 and 5 (son and mother) had 10/16 matches of HLA molecules. Moreover, subjects 6 and 7 (first cousins once removed) had 6/16 matches of HLA molecules (Table 2).
The HLA Typing Results in all the Subjects
| Subject | HLA-A | HLA-B | HLA-C | HLA-DR | HLA-DP | HLA-DQ |
|---|---|---|---|---|---|---|
| 1 | 11, 24 | 15, 40 | 04, 08 | B1 04, 12 | A1 01:03, 02:02; B1 02:02, 105:01 | A1 01, 03; B1 04, 05 |
| 2 | 11, 24 | 15, 51 | 08, 14 | B1 04, 14 | A1 02:01, 02:02; B1 05:01, 13:01 | A1 01, 03; B1 04, 05 |
| 3 | 11, 24 | 18, 18 | 07, 07 | B1 15, 15 | A1 02:02, 02:02; B1 02:02, 05:01 | A1 01, 01; B1 05, 06 |
| 4 | 11,33 | 38:02, 40 | 07, 08 | B1 12, 14 | A1 02:01, 02:02; B1 01:01, 13:01 | A1 01, 06:01; B1 03, 05 |
| 5 | 11, 24 | 39, 40 | 07, 08 | B1 04, 12 | A1 02, 02; B1 01:01, 03:01 | A1 03, 06:01; B1 03, 03 |
| 6 | 02:03, 11:01 | 15:139, 53:14 | 04:01, 08:01 | B1 15:02, 16:02 | A1 02,02: 02,02; B1 05:01, 05:01 | A1 01:01, 01:02; B1 05:01, 05:02 |
| 7 | 02:01, 11:01 | 18:01, 56:02 | 03:03, 07:04 | B1 14:04, 15:02 | A1 02:01, 02:02; B1 05:01, 13:01 | A1 01:01, 01:01; B1 05:01, 05:03 |
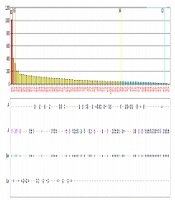
HLA-Abs to both class I and class II HLA molecules were found in subjects with no history of HLA exposure (subjects 1, 3, and 4), albeit with relatively low MFI values (Figure 1). Subject 6 had 59 HLA-Abs toward the both class I and class II HLA molecules with MFI values ranging from 1003.36-12404.77, none of which matched Subject 7’s HLA molecules (Table 3).
The HLA-Abs examination result toward class I (A) and class II (B) HLA in the subject never exposed to HLA molecules. Each bar corresponds to a specific HLA locus and allele, while the height of the bars corresponds to the MFI values. All of the HLAabs titer (MFI values) in this subject were < 1,500.
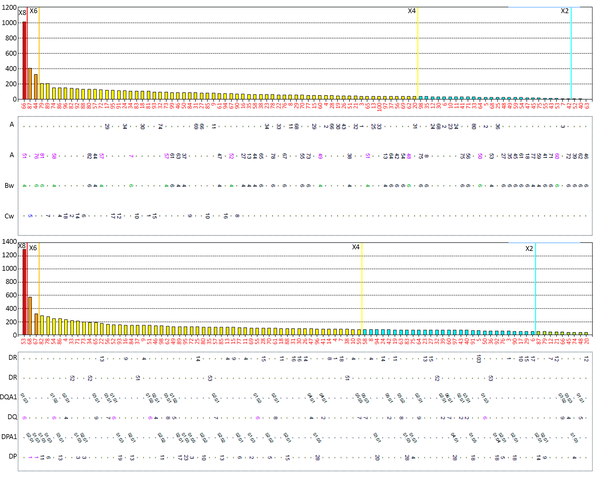
The HLA-Abs Examination Result in Subject 6
| HLA Class | HLA Locus | Allele | MFI |
|---|---|---|---|
| I | C 1 | C*01:02 | 3,775.99 |
| C 12 | C*12:03 | 3,484.48 | |
| C 15 | C*15:02 | 3,237.54 | |
| B 76 | B*15:12 | 1,003.79 | |
| B 63 | B*15:16 | 1,003.36 | |
| II | DR 14 | DRB1*14:54 | 12,404.77 |
| DRB1*14:01 | 10,864.22 | ||
| DRB1*14:02 | 3,338.05 | ||
| DR 9 | DRB1*09:01 | 10,870.63 | |
| DRB1*09:02 | 8,075.48 | ||
| DR 52 | DRB3*02:02 | 9,782.17 | |
| DRB3*03:01 | 3,107.66 | ||
| DR 1 | DRB1*01:01 | 8,974.78 | |
| DRB1*01:02 | 8,160.40 | ||
| DR 8 | DRB1*08:01 | 7,815,05 | |
| DR 7 | DRB1*07:01 | 6,702.68 | |
| DR 4 | DRB1*04:03 | 6,536.66 | |
| DRB1*04:04 | 5,267.12 | ||
| DRB1*04:05 | 3,991.46 | ||
| DRB1*04:02 | 3,896.42 | ||
| DRB1*04:01 | 3,685.12 | ||
| DR 103 | DRB1*01:03 | 6,139.51 | |
| DR 13 | DRB1*13:01 | 4,952.94 | |
| DRB1*13:03 | 4,159.30 | ||
| DR 12 | DRB1*12:02 | 4,683.66 | |
| DRB1*12:01 | 4,065.18 | ||
| DR 10 | DRB1*10:01 | 4,432.50 | |
| DR 17 | DRB1*03:01 | 3,955.12 | |
| DR 11 | DRB1*11:01 | 3,597.46 | |
| DRB1*11:04 | 2,896.87 | ||
| DR 18 | DRB1*03:02 | 2,936.17 | |
| DR 51 | DRB5*02:02 | 4,336.89 | |
| DRB5*01:01 | 3,457.45 | ||
| DR 52 | DRB3*01:01 | 3,265.07 | |
| DR 53 | DRB4*01:01 | 2,087.58 | |
| DR 15 | DRB1*15:01 | 1,005.29 | |
| DRB1*15:03 | 1,091.27 | ||
| DR 16 | DRB1*16:01 | 1,248.62 | |
| DR 53 | DRB4*01:03 | 1,285.38 | |
| DQ 2 | DQA1*05:01 | 8,463.38 | |
| DQB1*02:01 | 8,463.38 | ||
| DQA1*03:01 | 2,082.90 | ||
| DQB1*02:01 | 2,082.90 | ||
| DQA1*02:01 | 1,393.82 | ||
| DQB1*02:02 | 1,393.82 | ||
| DQA1*02:01 | 1,007.14 | ||
| DQB1*02:01 | 1,007.14 | ||
| DQ 7 | DQA1*05:03 | 1,581.14 | |
| DQB1*03:01 | 1,581.14 | ||
| DQA1*05:05 | 1,292.49 | ||
| DQB1*03:01 | 1,292.49 | ||
| DQ 8 | DQA1*03:02 | 1,411.45 | |
| DQB1*03:02 | 1,411.45 | ||
| DQ 9 | DQA1*03:02 | 1,367.01 | |
| DQB1*03:03 | 1,367.01 | ||
| DQA1*02:01 | 1,097.59 | ||
| DQB1*03:03 | 1,097.59 | ||
| DP 1 | DPA1*01:03 | 2,064.85 | |
| DPB1*01:01 | 2,064.85 |
The result was presented as the loci (HLA-A, -B, -C, -DR, -DP, and -DQ) and two alleles (one from each parent) for every locus. HLA-DP and -DQ had two sets of alleles for each locus: A1 and B1.
5. Discussion
This study found that subjects with a history of KT developed more HLA-Abs compared to those with no organ transplantation. This phenomenon is a consequence of less previous exposure to other HLA molecules, as proven with low HLA crossmatch results (4, 5). This strengthens previous knowledge that the immunologic aspect, mainly recipient-donor matching, is a vital step in the organ transplant process (4).
Since the first successful kidney allograft transplantation by Joseph Murray in 1954, the procedure has been the preferred mode of treatment for ESRD because it offers a more definitive outcome, i.e., longer survival and better quality of life, compared to dialysis treatment (6, 7). A systematic review of 110 eligible studies with a total 1,961,904 kidney failure patients comparing the clinical outcomes of KT and dialysis found that KT was associated with reduced risk of mortality and cardiovascular events as well as with better quality of life compared to treatment with dialysis. They suggested that the relative benefit of transplantation significantly increased over time compared to dialysis (8). However, as previously mentioned, rejection may occur following KT, which is mediated with HLA molecules (9).
HLA molecules are inherited through Mendelian principles with en-bloc HLA-A, -B, -C, -DR, and -DQ haplotype transmission from each parental chromosome, although recombination can occur within the HLA system (10). It means that 50% of HLA molecules in a person are derived from each of their parents and that siblings have 25% chance of having the same HLA molecules. Our result showed that the siblings and the son-mother pair had the highest number of HLA matches (10/16 matches each) compared to the other subjects. Subjects 6 and 7, who were first cousins once removed (one of Subject 7’ parents is Subject 6’ first cousin), had some similarity in their HLA although only had 6/16 HLA matches. Our result also showed that the same HLA loci were shared between many subjects, despite no familial relationship. This may suggest that there are common HLA molecules that may be race-related (4). This, however, needs further classification.
Analysis of HLA-Abs detection showed that prior exposure to HLA molecules, in this case, through previous kidney transplantation, triggered the development of HLA-Abs with high MFI values in subject 6. Currently, there is no specific threshold of MFI values for donor-recipient compatibility assessment in KT. The principle is to prevent graft kidney transplantation to a recipient with proven DSA(s) (11, 12). Although subject 6 had 59 different HLAabs, the HLA molecules that correspond to those antibodies were not present in subject 7 (donor candidate). KT was performed in subject 6 using the kidney of subject 7 as graft. Post-transplantation follow-up on subject 6 six months after the surgery still showed normal graft function.
The development of the SSOP method for HLA typing examination, which uses multiplexed beads (or microparticles) and flow cytometry to detect HLA molecules, yields more sensitive and specific results compared to other methods, such as complement-dependent cytotoxicity (CDC) examining B- and T-cells or conventional flow cytometry (13). The SSOP method provides a semi-quantitative numeric fluorescence value. In addition to its ability to perform HLA typing, this method is also capable of monitoring patient antibody profiles for particular specificities, such as the identification of de novo DSA post-transplantation (14). Although HLA-Abs can be identified with CDC, the SSOP method offers a number of benefits, including no requirement for viable lymphocytes and complement, detection of only HLA-specific antibodies, and the ability to detect non-complement fixing antibodies (14).
5.1. Conclusions
HLA typing and HLA-Abs detection for donor-recipient compatibility assessment using Luminex®-based assays offer more detailed, semi-quantitative results. However, it is still not clear whether HLA typing and HLA-Abs detection with the SSOP method are clinically superior to crossmatching with CDC. Furthermore, the clinical significance of pre-transplant, low-titer, and positive DSA in CDC-crossmatch negative kidney transplant recipients still remains inconclusive. Moreover, the DSA level associated with rejection is yet to be fully determined (13-15). Further studies using a sizeable number of samples in a cohort setting are needed to profile the MFI values at our center.
Acknowledgements
References
-
1.
Mahdi BM. A glow of HLA typing in organ transplantation. Clin Transl Med. 2013;2(1):6. [PubMed ID: 23432791]. [PubMed Central ID: PMC3598844]. https://doi.org/10.1186/2001-1326-2-6.
-
2.
Takemoto S, Port FK, Claas FH, Duquesnoy RJ. HLA matching for kidney transplantation. Hum Immunol. 2004;65(12):1489-505. [PubMed ID: 15603878]. https://doi.org/10.1016/j.humimm.2004.06.008.
-
3.
Konvalinka A, Tinckam K. Utility of HLA antibody testing in kidney transplantation. J Am Soc Nephrol. 2015;26(7):1489-502. [PubMed ID: 25804279]. [PubMed Central ID: PMC4483594]. https://doi.org/10.1681/ASN.2014080837.
-
4.
Tambur AR. Human leukocyte antigen matching in organ transplantation: What we know and how can we make it better (Revisiting the past, improving the future). Curr Opin Organ Transplant. 2018;23(4):470-6. [PubMed ID: 29750676]. https://doi.org/10.1097/MOT.0000000000000538.
-
5.
Cusick MF, Jindra PT. Human leukocyte antigen epitope matching in solid organ transplantation. Clin Lab Med. 2018;38(4):595-605. [PubMed ID: 30420055]. https://doi.org/10.1016/j.cll.2018.07.004.
-
6.
Tzvetanov I, D'Amico G, Benedetti E. Robotic-assisted kidney transplantation: Our experience and literature review. Curr Transplant Rep. 2015;2(2):122-6. [PubMed ID: 26000230]. [PubMed Central ID: PMC4431703]. https://doi.org/10.1007/s40472-015-0051-z.
-
7.
Merrill JP, Murray JE, Harrison JH, Guild WR. Successful homotransplantation of the human kidney between identical twins. J Am Med Assoc. 1956;160(4):277-82. [PubMed ID: 13278189]. https://doi.org/10.1001/jama.1956.02960390027008.
-
8.
Tonelli M, Wiebe N, Knoll G, Bello A, Browne S, Jadhav D, et al. Systematic review: Kidney transplantation compared with dialysis in clinically relevant outcomes. Am J Transplant. 2011;11(10):2093-109. [PubMed ID: 21883901]. https://doi.org/10.1111/j.1600-6143.2011.03686.x.
-
9.
Tuncer M, Gurkan A, Erdogan O, Yucetin L, Demirbas A. Lack of impact of human leukocyte antigen matching in living donor kidney transplantation: Experience at Akdeniz University. Transplant Proc. 2005;37(7):2969-72. [PubMed ID: 16213277]. https://doi.org/10.1016/j.transproceed.2005.07.043.
-
10.
Howell WM, Carter V, Clark B. The HLA system: Immunobiology, HLA typing, antibody screening and crossmatching techniques. J Clin Pathol. 2010;63(5):387-90. [PubMed ID: 20418230]. https://doi.org/10.1136/jcp.2009.072371.
-
11.
Tait BD, Susal C, Gebel HM, Nickerson PW, Zachary AA, Claas FH, et al. Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation. 2013;95(1):19-47. [PubMed ID: 23238534]. https://doi.org/10.1097/TP.0b013e31827a19cc.
-
12.
Karam G, Kälble T, Alcaraz A, Aki FT, Budde K, Humke U, et al. Guidelines on renal transplantation. Ren Transplant Eur Assoc Urol. 2009:327-37.
-
13.
Wu P, Jin J, Everly MJ, Lin C, Terasaki PI, Chen J. Impact of alloantibody strength in crossmatch negative DSA positive kidney transplantation. Clin Biochem. 2013;46(15):1389-93. [PubMed ID: 23726814]. https://doi.org/10.1016/j.clinbiochem.2013.05.053.
-
14.
Bshi/Bts. Guidelines for the detection and characterisation of clinically relevant antibodies in allotransplantation. 2014. 21 p.
-
15.
Mohan S, Palanisamy A, Tsapepas D, Tanriover B, Crew RJ, Dube G, et al. Donor-specific antibodies adversely affect kidney allograft outcomes. J Am Soc Nephrol. 2012;23(12):2061-71. [PubMed ID: 23160511]. [PubMed Central ID: PMC3507372]. https://doi.org/10.1681/ASN.2012070664.