Abstract
Background:
Nosocomial infections are important medical concerns in developed and developing countries. Pseudomonas aeruginosa is considered the third leading cause of nosocomial infections following Staphylococcus aureus and Escherichia coli.Objectives:
The present study aimed to determine the antibiotic resistance of isolated P. aeruginosa strains in the patients admitted to Imam Reza Hospital in Kermanshah, Iran.Methods:
This descriptive, cross-sectional study was conducted on 900 patients with positive P. aeruginosa cultures who were admitted to Imam Reza Hospital in Kermanshah, Iran during 2016-2018. Bacterial isolates were separated using laboratory tests, and the standard disc-diffusion method was used to assess antibiotic susceptibility based on the CLSI protocol. Data analysis was performed in SPSS version 24.Results:
The majority of the P. aeruginosa-positive cases were isolated from the emergency ward (44.3%), intensive care unit (21.9%), blood samples (40.4%), and urine samples (18.7%). The highest antibiotic resistance was observed against cotrimoxazole, ceftriaxone, ampicillin, ampicillin-sulbactam, nitrofurantoin, nalidixic acid, cefazolin, and cefixime. In addition, ciprofloxacin and imipenem were the most effective antibiotics against P. aeruginosa with the sensitivity of 68.1% and 57.2%, respectively.Conclusions:
According to the results, P. aeruginosa had high resistance against antibiotics such as cotrimoxazole and ceftriaxone. Therefore, these antibiotics should be used correctly and reasonably, and epidemiological studies in this regard should be focused on proposing national programs to prevent the further spread of antibiotic resistance.Keywords
Nosocomial Infection Pseudomonas aeruginosa Antibiotic Resistance
1. Background
Hospitalized patients may be at the risk of nosocomial bacteria, which cause secondary infections and hinder the recovery process (1). The treatment of nosocomial infections is also associated with the development of antibiotic resistance in the patients. Pseudomonas aeruginosa has been identified in critically ill patients as part of the normal intestinal flora, as well as an important pathogen responsible for multiple intensive care unit (ICU) infections (2). The establishment rate of P. aeruginosa is on the rise in patients with a long hospitalization period (3). On the other hand, most of these bacterial strains have become resistant to various antibiotics and are classified as multidrug-resistant strains (3, 4).
P. aeruginosa has multiple virulence factors, which could abnormally reduce the neutrophils in immature individuals and patients with cystic fibrosis, abnormal neutrophil depletion, and various systemic infections. The infections caused by this microorganism occur both in the population and in hospitals, while the prevalence in hospital environments contributes to other chronic infections, especially in patients with the impaired immune system and cystic fibrosis (5).
The most commonly used agents in the treatment of P. aeruginosa infections include broad-spectrum penicillins (carbenicillin, ticarcillin, and piperacillin), broad-spectrum cephalosporins (ceftazidime and cefepime), carbapenems, aminoglycosides, fluoroquinolones, and aztreonam (3, 4). Furthermore, wide-spectrum beta-lactamase enzymes (ESBLs) could mediate the resistance of P. aeruginosa strains to second- and third-generation (broad-spectrum) cephalosporins and monobactams. Various genes have also been detected on the chromosome or plasmid (6, 7), which encode these enzymes (8).
Several pathways are involved in the antibiotic resistance of P. aeruginosa, with the innate resistance encompassing over-expressed efflux systems and the relatively low membrane permeability (6). Acquired resistance also involves the acquisition of resistance genes or mutations in the genes encoding porins, efflux pumps, penicillin-binding proteins, and chromosomal b-lactamase, which lead to resistance to b-lactams, carbapenems, aminoglycosides, and fluoroquinolones (7).
2. Objectives
A review of the literature shows that the resistance pattern of P. aeruginosa to various antibiotics is constantly changing depending on the countries using antibiotics and type of the strains; therefore, such investigations should continue in across the world. The present study aimed to determine the frequency and pattern of antibiotic resistance in various wards of Imam Reza (AS) Hospital in Kermanshah, Iran during 2016-2018.
3. Methods
In this descriptive, cross-sectional study, a laboratory expert provided the list of the patients admitted to Imam Reza Hospital during 2016-2018 with positive P. aeruginosa cultures in the blood, urine, wound, pleura, and cerebrospinal fluid samples. The susceptibility pattern and antibiotic resistance of the pseudomonas isolated from each patient were reported. Following that, the samples were sent to the microbiology laboratory, grown in eosin methylene blue and blood agar, and incubated at the temperature of 37°C for 24 hours. After colony growth and the initial identification of the organism, the bacterium was identified based on the colony color, presence/absence of hemolysis, and gram-smear staining prepared from the colonies.
At the next stage, antibiogram was performed using the disc-diffusion method, and the exact diameter of the growth inhibition zone was determined in millimeters using the Müller-Hinton agar medium. In addition, comparison was performed with the Clinical and Laboratory Standard Institute (CLSI) standard table for gentamicin (10 µg), ciprofloxacin (5 µg), ceftazidime (30 µg), cotrimoxazole (1.25 µg), ceftriaxone (30 µg), cefepime, imipenem, ampicillin (10 µg), piperacillin (100 µg), ampicillin-sulbactam (10 µg), nitrofurantoin (300 µg), nalidixic acid (30 µg), cefixime (5 µg), levofloxacin (5 µg), and tazobactam (110 µg).
The bacterial suspension was prepared with the turbidity equivalent to 0.5 McFarland of the tube turbidity (× 108 CFU/ml 1.5), and lawn culture was performed in triplicate using a sterile swab and a plate containing the Müller-Hinton agar medium. In addition, pliers were used to remove the discs from the freezer one hour before they were placed on the culture medium and stabilized with the tip of a pair of pliers; following that, the plates were incubated at the temperature of 37°C for 24 hours. To read the results, the diameter of the inhibition zone was measured in millimeters using an accurate ruler, and the obtained results were reported as sensitivity (S1), resistance (R2), and semi-sensitive zones (I3).
3.1. Ethical Considerations
The study protocol was ethically approved, and the confidentiality of information was also observed.
3.2. Statistical Analysis
Data analysis was performed in SPSS version 18 using descriptive statistics (mean, standard deviation, frequency, and percentage). In addition, Chi-square (X2) and independent t-test were applied to evaluate the correlations between antibiotic resistance, infection prevalence, and infection severity. In all the statistical analyses, P value of 0.05 was considered significant.
4. Results
In 2016, 2017, and 2018, 213 (23.7%), 221 (24.6%) and 466 positive cases of pseudomonas (51.8%) were reported among the patients admitted to Imam Reza Hospital. Out of 900 patients with P. aeruginosa positive cultures in the mentioned periods, 61.6% were male, and 38.4% were female. Among the positive culture samples, 28.7% were urine samples, 18.7% were sputum samples, 40.4% were blood samples, 1.2% were eye samples, 5,4 % were wound cultures, 5.3% were fluid samples, and 0.1% was accounted for by vaginal and stomach samples. Table 1 shows the number of the positive cases of P. aeruginosa infection by hospital sections. The antibiogram is presented in Table 2 and Figure 1.
Positive Cases of Pseudomonas aeruginosa by Wards of Imam Reza Hospital (2016-2018)
| Hospital Wards | Percentage, % |
|---|---|
| ICUs (general, neonatal, pediatric) | 19.7 |
| Emergency Department | 44.3 |
| Internal (internal, infections, pediatric, neurology) | 19 |
| Surgery (general, gynecological, urology) | 12.1 |
| Outpatients | 2 |
| Transplantation (bone marrow, kidney) | 2.9 |
| Total | 100 |
Antibiogram of P. aeruginosa in Positive Cultures (2016-2018)
| Antibiotic | Sensitive (%) | Semi-sensitive (%) | Resistant (%) |
|---|---|---|---|
| Ciprofloxacin | 68.1 | 5.5 | 26.3 |
| Ceftazidime | 44.2 | 11.9 | 43.9 |
| Cotrimoxazole | 16.3 | 1.5 | 82.1 |
| Ceftriaxone | 18 | 13.7 | 68.3 |
| Cefepime | 25.5 | 21.2 | 53.3 |
| Gentamicin | 55.9 | 12.4 | 31.7 |
| Imipenem | 57.2 | 8.4 | 34.4 |
| Ampicillin | 20.4 | 3.9 | 75.7 |
| Piperacillin | 43 | 10.7 | 46.3 |
| Ampicillin-Sulbactam | 13.5 | 1.6 | 84.9 |
| Nitrofurantoin | 4.8 | 3.9 | 91.3 |
| Nalidixic Acid | 2.5 | 5.5 | 92 |
| Levofloxacin | 36.4 | 27.3 | 36.4 |
| Tobramycin | 42.9 | 14.3 | 42.9 |
| Tazobactam | 42.9 | 10.7 | 46.4 |
| Cefazolin | 1.2 | 1.8 | 97 |
| Amikacin | 36.2 | 25 | 38.8 |
| Cefotaxime | 13.9 | 19 | 67.1 |
| Cefixime | 7.1 | 8.6 | 84.3 |
P. aeruginosa Resistance to Various Antibiotics in Imam Reza Hospital (2016-2018)
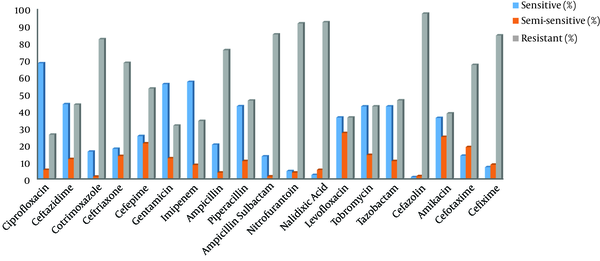
The highest antibiotic resistance was observed against cefazolin, nalidixic acid, nitrofurantoin, ampicillin /sulbactam, and cotrimoxazole. On the other hand, ciprofloxacin and imipenem were the most effective antibiotics against P. aeruginosa infection with the sensitivity of 68.1% and 57.2%, respectively.
5. Discussion
P. aeruginosa is considered to be the most common pathogen to cause nosocomial infections, as well as the leading cause of nosocomial respiratory tract infections (9). The active presence of P. aeruginosa in a wide range of hospitalized patients could be attributed to the unique features of this bacterium. In the current research, P. aeruginosa was detected in the patients admitted to different hospital wards and in blood and urine samples, which indicated the role of this bacterium in urinary tract infections as well.
Our findings based on the spectrum of P. aeruginosa are consistent with the study by Tavajohi et al. (10), which demonstrated that the prevalence of P. aeruginosa was highest in urine and blood culture samples. According to the results of the present study, antibiotic resistance was higher in the emergency department and ICU compared to the other wards. In ICUs, factors such as medical restrictions, impairment due to long hospital stay, and use of multiple care equipment may contribute to the increased prevalence of P. aeruginosa-resistant strains.
The current research indicated that the highest antibiotic resistance against cefazolin, nalidixic acid, nitrofurantoin, ampicillin-sulbactam, cefixime, and cotrimoxazole. In the study conducted by Oulia et al. (11) on 100 isolates of P. aeruginosa, the resistance of the strains to cefazolin, cephalexin, ceftriaxone, ceftizoxime, cefixime, and ciprofloxacin was reported to be 100%, 100%, 92%, 94%, 100%, and 89%, respectively, which is consistent with the results of the present study in terms of bacterial resistance to cefazolin and cefixime. In this regard, Abdi et al. (12) reported ampicillin-sulbactam to be one of the most resistant antibiotics in the treatment of P. aeruginosa infections, which is also in line with the results of the present study.
In a descriptive analysis conducted in Kashan (Iran), Tavajohi et al. (10) reported that P. aeruginosa isolates showed the highest resistance respectively against piperacillin, imipenem, cefotaxime, ceftriaxone, gentamicin, ceftazidime, aztreonam, and ciprofloxacin. In another descriptive research, Salehi et al. (2015) (13) observed that 100% of bacterial isolates were resistant to nalidixic acid, and 96% were resistant to norfloxacin and ciprofloxacin. Based on the aforementioned studies, it could be concluded that the increased resistance of bacterial strains to fluoroquinolones is due to various factors, such as the indiscriminate use of antibiotics in clinical centers. In the present study, imipenem was the most effective antibiotic after ciprofloxacin. In the study by Rajabpour et al. (14), the highest resistance was observed against ciprofloxacin (58%) and levofloxacin (61.2%), while the lowest resistance was against imipenem (9.6%) among eight selected antibodies.
In the current research, ciprofloxacin resistance in the isolates was estimated at 26.3%, while it has been reported to be 26.8% in Latin America (15) and 10-32% in Europe (16-18). In some studies, ciprofloxacin resistance has been higher compared to our findings. For instance, the research by Behera et al. (19) indicated resistance to this antibiotic to be more than 75%, while Rubin et al. (20) reported the resistance rate of 39% to ceftriaxone, which is lower than our findings. In the present study, cefotaxime showed 67.1% resistance, which is higher than the reported rate by Rustini et al. (21).
5.1. Conclusion
According to the results, the increased resistance of P. aeruginosa strains to a wide range of antibiotics in treatment centers has become a major clinical issue, especially in burns, lungs, and ICU wards. Due to high genetic diversity, it is not possible to definitively determine the antibiotics that are effective in the treatment of P. aeruginosa infections in various patients as there are differences even within a small geographical area. Therefore, the determination of the most effective drugs for the treatment of these infections depends on the antibiogram test of the isolated samples from each patient independently. We observed no significant difference in drug resistance between the proposed and non-recommended CLSI antibiotics. Due to the high antibiotic resistance and the fact that the treatment of nosocomial infections is difficult and may lead to the death of the patients or threaten all hospitalized patients, most nosocomial infections should be controlled to prevent their spread with lower costs by using microbiological diagnostic methods.
Acknowledgements
References
-
1.
Wong YP, Chua KH, Thong KL. One-step species-specific high resolution melting analysis for nosocomial bacteria detection. J Microbiol Methods. 2014;107:133-7. [PubMed ID: 25307691]. https://doi.org/10.1016/j.mimet.2014.10.001.
-
2.
Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6(2):109-19. [PubMed ID: 31194018]. [PubMed Central ID: PMC6545445]. https://doi.org/10.1016/j.gendis.2019.04.001.
-
3.
Rezaee MA, Pajand O, Nahaei MR, Mahdian R, Aghazadeh M, Ghojazadeh M, et al. Prevalence of Ambler class A beta-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2013;76(3):330-4. [PubMed ID: 23726148]. https://doi.org/10.1016/j.diagmicrobio.2013.04.003.
-
4.
Rumbo C, Gato E, Lopez M, Ruiz de Alegria C, Fernandez-Cuenca F, Martinez-Martinez L, et al. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57(11):5247-57. [PubMed ID: 23939894]. [PubMed Central ID: PMC3811325]. https://doi.org/10.1128/AAC.00730-13.
-
5.
Gomes MZ, Oliveira RV, Machado CR, Conceicao Mde S, Souza CV, Lourenco MC, et al. Factors associated with epidemic multiresistant Pseudomonas aeruginosa infections in a hospital with AIDS-predominant admissions. Braz J Infect Dis. 2012;16(3):219-25. [PubMed ID: 22729187]. https://doi.org/10.1590/s1413-86702012000300001.
-
6.
Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int. 2016;2016:2475067. [PubMed ID: 27274985]. [PubMed Central ID: PMC4871955]. https://doi.org/10.1155/2016/2475067.
-
7.
Oie S, Fukui Y, Yamamoto M, Masuda Y, Kamiya A. In vitro antimicrobial effects of aztreonam, colistin, and the 3-drug combination of aztreonam, ceftazidime and amikacin on metallo-beta-lactamase-producing Pseudomonas aeruginosa. BMC Infect Dis. 2009;9:123. [PubMed ID: 19664245]. [PubMed Central ID: PMC2738676]. https://doi.org/10.1186/1471-2334-9-123.
-
8.
Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2010;36(1):70-4. [PubMed ID: 19524369]. https://doi.org/10.1016/j.burns.2009.01.015.
-
9.
Jarvis WR, Martone WJ. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29 Suppl A:19-24. [PubMed ID: 1601752]. https://doi.org/10.1093/jac/29.suppl_a.19.
-
10.
Tavajjohi Z, Moniri R, Khorshidi A. Frequency of extended-spectrum beta-lactamase (ESBL) multidrug-resistance produced by Pseudomonas aeruginosa isolated from clinical and environmental specimens in Kashan Shahid Beheshti hospital during 2010-11. Feyz J Kashan Univ Med Sci. 2011;15(2).
-
11.
OULIA P, NAMAYAN B, SADERI H, MANSOURI S. COMPARISON OF THE EFFECTS OF CIPROFLOXACIN & CEPHALOSPORINS ON STRAINS OF PSEUDOMONAS AERUGINOSA ISOLATED FROM BURN WOUND INFECTIONS & DETERMINATION OF MIC OF GENTAMICIN. Daneshvar Med. 2004;11(50):1-6.
-
12.
Abdi M, Soltan Dallal MM, Hajiabdolbaghi M, Douraghi M, Davoodabadi A. Study of antagonistic properties of lactobacilli isolated from healthy baby stools on growth of Acinetobacter baumannii and Pseudomonas aeruginosa of nosocomial origin. Pajoohandeh J. 2016;20(6):302-7.
-
13.
Salehi M, Samani S. Determining the Prevalence of qnrB Gene in Pseudomonas Aeruginosa Strains Isolated from Clinical Centers in Tehran in 1394. Alborz Univ Med J. 2018;7(3):190-6. https://doi.org/10.29252/aums.7.3.190.
-
14.
Rajabpour M, Alikhani MY. MIC determination of Pseudomonas aeruginosa strains were isolated from clinical specimens of patients admitted to educational hospitals in Hamedan (90-91). Iranian J Med Microbiol. 2013;7(3):18-25.
-
15.
Jones RN. Resistance patterns among nosocomial pathogens: trends over the past few years. Chest. 2001;119(2 Suppl):397S-404S. [PubMed ID: 11171776]. https://doi.org/10.1378/chest.119.2_suppl.397s.
-
16.
Bonfiglio G, Carciotto V, Russo G, Stefani S, Schito GC, Debbia E, et al. Antibiotic resistance in Pseudomonas aeruginosa: an Italian survey. J Antimicrob Chemother. 1998;41(2):307-10. [PubMed ID: 9533479]. https://doi.org/10.1093/jac/41.2.307.
-
17.
Bouza E, Garcia-Garrote F, Cercenado E, Marin M, Diaz MS. Pseudomonas aeruginosa: a survey of resistance in 136 hospitals in Spain. The Spanish Pseudomonas aeruginosa Study Group. Antimicrob Agents Chemother. 1999;43(4):981-2. [PubMed ID: 10103217]. [PubMed Central ID: PMC89243]. https://doi.org/10.1128/AAC.43.4.981.
-
18.
Spencer RC, Clark JC. Prophylaxis after splenectomy. J Antimicrob Chemother. 1994;34(2):295. [PubMed ID: 7814293]. https://doi.org/10.1093/jac/34.2.295.
-
19.
Behera B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-beta-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26(3):233-7. [PubMed ID: 18695320]. https://doi.org/10.4103/0255-0857.39587.
-
20.
Rubin J, Walker RD, Blickenstaff K, Bodeis-Jones S, Zhao S. Antimicrobial resistance and genetic characterization of fluoroquinolone resistance of Pseudomonas aeruginosa isolated from canine infections. Vet Microbiol. 2008;131(1-2):164-72. [PubMed ID: 18395369]. https://doi.org/10.1016/j.vetmic.2008.02.018.
-
21.
Rustini R, Jamsari J, Marlina M, Zubir N, Yuliandra Y. Antibacterial Resistance Pattern of Pseudomonas Aeruginosa Isolated from Clinical Samples at a General Hospital in Padang, West Sumatra, Indonesia. Asian J Pharm Clin Res. 2017;10(8):158. https://doi.org/10.22159/ajpcr.2017.v10i8.18539.