Abstract
Background:
Acinetobacter baumannii is one of the major bacteria causing nosocomial infections. The emergence of multidrug-resistant (MDR) isolates causes morbidity and mortality and lead to financial burdens for patients and public health systems.Objectives:
Regarding the use of ciprofloxacin and increase of ciprofloxacin resistance, this study was aimed to investigate the role of the efflux pump and mutations in gyrA and parC genes as the mechanisms of ciprofloxacin-resistance.Methods:
This study was conducted on 55 strains of A. baumannii isolated from the patients hospitalized in Milad Hospital, Tehran. The isolates were identified by biochemical tests, and their antibiotic susceptibility was assessed by the disc diffusion method. To investigate the role of the efflux pump, minimum inhibitory concentration (MIC) of ciprofloxacin was determined in the presence and absence of the carbonyl cyanide m-chlorophenyl hydrazone (CCCP) inhibitor. The PCR was used for amplification of gyrA, and parC genes and sequencing of its products was carried out to track the mutations.Results:
The highest and lowest antibiotic resistance were observed in ciprofloxacin (100%) and tobramycin (52.7%), respectively. Moreover, 96% of the A. baumannii isolates exhibited MDR. Five percent of strains showed 4-fold MIC reduction after the use of the inhibitor and were reported as the strains with high activity efflux pump. In 100% of the isolates, gyrA and parC genes were detected, and only one mutation was observed in parC and gyrA genes in the studied isolates that altered amino acid type.Conclusions:
Reduction in MIC in the presence of an efflux pump inhibitor confirmed its role in enhancing the resistance to ciprofloxacin in some isolates. The results of this study also revealed no significant relationship between the ciprofloxacin resistance of the studied isolates and mutation in the encoding region of parC and gyrA genes. Thus, the significance of other mechanisms, such as the expression of genes encoding efflux pump proteins, should not be neglected.Keywords
Acinetobacter baumannii Ciprofloxacin Resistance Efflux Pump QRDRs Mutation
1. Background
Among Acinetobacter species, Acinetobacter baumannii is the most common human-isolated species (1). These organisms are widely spread in nature and can be found in soil, water, and wastewater (2). Acinetobacter baumannii is the major cause of a wide range of nosocomial infections such as blood infection, ventilator-related pneumonia, urinary tract infections, and ulcer infections (3, 4). Single cases of peritonitis, endocarditis, meningitis, osteomyelitis, and arthritis have also been reported regarding A. baumannii-induced infections (5, 6). Some potential virulence factors of A. baumannii seem to be important for disease, including outer membrane porins, surface structures, e.g., capsule and lipopolysaccharide, enzymes such as phospholipase D, iron acquisition systems, and regulatory proteins (7). Partial treatment and mortalities due to A. baumannii-induced infections are among the major challenges of the medical communities (1).
Resistance to antibiotics and the emergence of multidrug-resistant (MDR) isolates are among the important causes of failure treatment of A. baumannii infections (5, 8). Studies showed that more than 80% of A. baumannii isolates are resistant to aminoglycosides or quinolones, such as ciprofloxacin and levofloxacin, and currently carbapenems have been extensively reported (9). However, A. baumannii has increasingly become resistant to these antibiotics and is only susceptible to the polymyxin antibiotic (10, 11). The main mechanisms for multiple drug resistant (MDR) of A. baumannii include horizontal gene transfer, increased expression of β-lactamases, alterations of membrane permeability, and increased expression of efflux pumps and genes encoding the target enzymes (12, 13). Bacterial efflux systems collect different compounds, such as antibiotics, from the cell and exhaust them; hence, they reduce the accumulation of antibiotics and increase their minimum inhibitory concentration (MIC) (14, 15). The bacterial efflux pumps belong to five large families, among which the Resistance Nodulation Division (RND) pump is a multidrug and three-component pump that uses ATP energy for substrate transfer (16, 17).
Inhibitors have been employed to inhibit the efflux pumps in bacteria; in this regard, carbonyl cyanide 3-chlorophenyl hydrazone (CCCP) is one of the most important inhibitors (15). This inhibitor belongs to the RND family and disrupts the function of the efflux pump; hence, it increases the accumulation of antibiotics within the bacterial cell, which in turn increases the drug efficacy (15). Another mechanism of resistance to ciprofloxacin involves the mutation in Quinolone Resistance Determining Regions (QRDRs), where target enzymes such as DNA gyrase and IV topoisomerase are affected (18). A mutation in the DNA gyrAse enzyme-encoding gene will inhibit the transcription process (18). DNA gyrase is a tetrameric enzyme with two subunits A and two subunits B encoded by gyrA and gyrB genes, respectively. They open up the negative coiling of the DNA. Topoisomerase IV is also a tetrameric enzyme consisting of two C and two E subunits encoded by parC and parE genes, respectively. They contribute to separating a female chromosome in genome replication (19, 20).
2. Objectives
Regarding the significance of ciprofloxacin in treating A. baumannii-induced infections and determining the mechanisms of antibiotic resistance, the present study was designed to investigate the resistance mechanisms such as the role of the efflux pumps and mutations in gyrA and ParC genes in the clinically isolated ciprofloxacin-resistant A. baumannii strains.
3. Methods
3.1. Sampling and Isolation of Acinetobacter baumannii Isolates
This descriptive cross-sectional study was conducted on A. baumannii strains isolated from the clinical samples (including the ulcers, blood, and urine) of 230 patients hospitalized in the intensive care unit (ICU) of Milad Hospital in Tehran, Iran from November 2017 to April 2018. Clinical samples were cultured on blood Agar and MacConkey agar to isolate A. baumannii strains. After 24 - 48 hours of incubation, they were identified by Gram staining and standard biochemical tests such as catalase, oxidase, culture in TSI (Merck, Germany), MRVP (Merck, Germany) and Urea (Merck, Germany) media and OF, Lysine, and bile esculin tests as well as growth at 42°C.
3.2. Antibiotic Susceptibility and Determination of MDR Strains
Antibiotic susceptibility of A. baumannii strains was assessed using disk diffusion method according to CLSI 2018. Antibiotic disks were supplied from Padtan Teb Company (Iran) containing gentamicin (10 μg), ciprofloxacin (5 μg), Ticarcillin (75 μg), doxycycline (30 μg), tobramycin (10 μg), levofloxacin (5 μg), imipenem (10 μg), and meropenem (10 μg). Bacterial concentration based on 0.5 McFarland standard was 1.5 × 108 CFU/mL. The results were reported as resistant, intermediate, and susceptible (Table 1). The A. baumannii (ATCC19606) was employed as the quality control strain. Strains showing resistance to more than 2 antibiotic classes were recognized as MDR strains.
Antibiotics and Their Growth Inhibition Zone Based on CLSI 2018
| Antibiotic | Disk Content (µg) | Sensitive (S) | Intermediate (I) | Resistant (R) |
|---|---|---|---|---|
| Ciprofloxacin | 5 | ≥ 21 | 16 - 20 | ≤ 15 |
| Ticarcillin | 75 | ≥ 20 | 15 - 19 | ≤ 14 |
| Tobramycin | 10 | ≥ 15 | 13 - 14 | ≤ 12 |
| Levofloxacin | 5 | ≥ 17 | 14 - 16 | ≤ 13 |
| Doxycycline | 30 | ≥ 13 | 10 - 12 | ≤ 9 |
| Imipenem | 10 | ≥ 22 | 19 - 21 | ≤ 18 |
| Gentamycin | 10 | ≥ 15 | 13 - 14 | ≤ 12 |
| Meropenem | 10 | ≥ 18 | 15 - 17 | ≤ 14 |
3.3. Efflux Pump Activity Assay
The minimum inhibitory concentration (MIC) of ciprofloxacin was determined by broth micro-dilution method based on CLSI 2018 in 96-well plates. Ciprofloxacin powder was dissolved in sterilized deionized water or a suitable solvent according to the manufacturers' instruction. Then, different concentrations (256, 128, 64, 32, 16, 8, 4, 2, 0.5, and 0.25 μg/mL) were prepared. Subsequently, 100 μL of the as-prepared dilutions was added to each well of the 96-well micro-plate. The concentration of the microbial suspension was set to 0.5 McFarland, and was diluted at the ratio of 1/100. Then, 100 μL of the microbial suspension was also added to each well, and the plates were incubated at 35°C for 18 - 24 hours. The lowest antibiotic concentration showing no growth was determined as MIC 90 (21).
To investigate the efflux pump activity, CCCP inhibitor with a final concentration of 25 μg/mL was added to each Muller Hinton Agar plates containing 0.25 - 256 μg/mL of ciprofloxacin. Once again, the minimum inhibitory concentration of antibiotics was determined after CCCP treatment. The 4-fold reduction in the MIC after the CCCP inhibitor application was considered an indicator of severe efflux pump activity. All the experiments were performed in 3 replicates. A CCCP-containing and antibiotic-free plate was applied as the control sample.
3.4. Molecular Analysis
3.4.1. Replication of gyrA and parC Genes by PCR
The Gram-negative bacteria extraction kit (Sinacolon Co., Iran) was employed for DNA extraction of all A. baumannii isolates. The quality and quantity of the extracted DNA were evaluated by gel agarose electrophoresis and NanoDrop spectrophotometry method at two wavelengths of 260 and 280 nm. A 260/280 ratio of ~1.8 is generally accepted as “pure” for DNA. To replicate and assess the frequency of gyrA genes, the specific primer sequence of gyrA-F (5'-AAATCTGCTCGTGTCGTTGG-3') and gyrA-R (5'-GCCATACCTACAGCAATACC-3') primer sequences with 349 bp size were applied; while the ParC-F (5'-AAGCCCGTACAGCGC CGTATT-3') and ParC-R (5'-AAAGTTATCTTGCCATTCGCT-3') primer sequence with 327 bp size were used to replicate (17).
For PCR reaction, a PCR mixture was used that contained 12.5 µL of 2× Master Mix (Sinacolon Co., Iran), including 1× PCR buffer, 1.5 mmol/L MgCl2, dNTPs at a concentration of 0.15 mmol/L each dNTP, 1.25 U of Taq DNA polymerase, 0.5 µL of 0.8 µM of each primer, 1 µL of template DNA (0.5 µg), and sterile distilled water up to 25 µL. The PCR temperature and time schedule involved the initial denaturation stage at 94°C for 5 minutes, followed by 55 seconds of denaturation at 94°C, the annealing phase at 54°C for 55 seconds, 32 cycles of 1-minute expansion stage at 72°C, and the final extension stage at 72°C for 5 minutes. To evaluate the PCR products, the samples were transferred to a 2% agarose gel; they were analyzed after staining in the Gel Doc apparatus (Vilber Lourmat, France). The positive control was A. baumannii ATCC 19606. The negative control contained no template.
3.4.2. Sequencing of PCR Products
To investigate the mutations in nucleotide sequences, the PCR products of a number of samples with confirmed ciprofloxacin resistance were purified and diluted in the proper concentration (1 µg/mL). The PCR amplified the purified sequences with both forward and reverse primers using BigDye technology by Bioneer Company (Germany). The sequencing results of PCR products were analyzed at the NCBI site (https://www.ncbi.nlm.nih.gov) using the BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to investigate the mutations in nucleotide sequences.
3.5. Statistical Data Analysis
SPSS version 20 (SPSS, Inc., Chicago, IL, USA) was employed for statistical analysis. Descriptive statistics and Pearson's chi‐square tests were used to evaluate the correlation between mutation and ciprofloxacin resistance. Statistical significance was defined as P value of less than 0.05.
4. Results
Fifty-five strains of A. baumannii were identified and isolated from clinical samples of 230 patients.
4.1. Antibiotic Resistance Pattern of Acinetobacter baumannii Isolates
The highest antibiotic resistance was observed in ciprofloxacin (100%), meropenem (96.4%), ticarcillin (94.6%) and imipenem (94.6%), respectively. Doxycycline (65.5%), gentamicin (58.2%), and tobramycin (52.7%) indicated the lowest resistance, respectively (Table 2). The results indicated that 96.36% of the isolates were MDR.
| Antibiotic | Resistant (R) | Intermediate (I) | Susceptible (S) |
|---|---|---|---|
| Ciprofloxacin | 100 | 0 | 0 |
| Ticarcillin | 94.6 | 1.8 | 3.6 |
| Tobramycin | 52.7 | 5.5 | 41.8 |
| Levofloxacin | 63.6 | 20 | 16.4 |
| Doxycycline | 65.5 | 3.6 | 30.9 |
| Imipenem | 94.6 | 1.8 | 3.6 |
| Gentamycin | 58.2 | 3.6 | 38.2 |
| Meropenem | 96.4 | 0 | 3.6 |
4.2. MIC of Ciprofloxacin
Investigation of ciprofloxacin MIC in A. baumannii strains indicated that 100% of the samples exhibited high resistance to this antibiotic (MIC ≥ 16 μg/mL).
4.3. Efflux Pump Activity After Application of CCCP
After employing CCCP inhibitor and determining the MIC of ciprofloxacin in the presence of this inhibitor, A. baumannii strains showing at least a 4-fold decrease in their ciprofloxacin MIC were phenotypically considered the strains with high efflux pump activity (Table 3). The results revealed that, among the ciprofloxacin-resistant MDR isolates, 3 strains (5%) exhibited a 4-fold decrease in their ciprofloxacin MIC in the presence of efflux pump inhibitor. Therefore, they were defined as the high efflux pump-active strains, while among 52 strains (26%) did not have efflux pump activity as they did not show any changes in their MIC, and 47% isolates showed that their MIC reduction was less than 4-fold (Figure 1).
Ciprofloxacin MIC of Some Strains Before and After Using CCCP Inhibitor and Determination of the Efflux Pump Activity
| Strain code | Ciprofloxacin MIC (µg/mL) | MIC Ciprofloxacin After Using CCCP (µg/mL) | MIC Reduction Rate | Efflux Pump Activity |
|---|---|---|---|---|
| 1 | 128 | 128 | - | - |
| 2 | 256 | 128 | 2 | - |
| 3 | 64 | 64 | - | - |
| 4 | 256 | 128 | 2 | - |
| 5 | 512 | 128 | 4 | + |
| 9 | 512 | 256 | 2 | - |
| 14 | 128 | 32 | 4 | + |
| 15 | 32 | 16 | 2 | - |
| 16 | 64 | 64 | - | - |
| 18 | 128 | 64 | 2 | - |
| 33 | 512 | 128 | 4 | + |
Frequency of high efflux pump activity in ciprofloxacin-resistant Acinetobacter baumannii strains after using CCCP is shown

4.4. Molecular Analysis Results
4.4.1. Determining the Frequency of gyrA and parC Genes by PCR Method
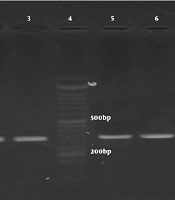
After replication of gyrA and parC genes, the PCR products of all A. baumannii strains were observed in form of bands with the length of gyrA 349bp and parC 328bp on the agarose gel (Figure 2). The results indicated that all of the studied strains that were ciprofloxacin-resistant (55 strains) because they possessed gyrA and parC genes.
Gel electrophoresis of PCR products. Lanes 1 - 3: bands for parC gene (327 bp), lanes 5 - 8: bands for ParC gene (349 bp), and lane 4: ladder (50 bp).

4.4.2. Mutation in gyrA and parC Genes
The study of A. baumannii resistant to ciprofloxacin with MIC ≥ 4µg/mL showed a mutation in parC gene in the studied samples and this sequence has the number 84 position mutation L > S. Among the sequenced A. baumannii isolates, which were resistant to ciprofloxacin with MIC ≥ 4 µg/mL considering gyrA gene, only one case had a mutation with the number 345 position mutation T > C, but the mutation did not alter the amino acid. There is no significant relationship between gene mutation and ciprofloxacin resistance among studied A. baumannii isolates (P > 0.05).
5. Discussion
Acinetobacter baumannii is the third cause of nosocomial pneumonia and ninth cause of blood infection in the hospitals (21, 22). Following the increase in the use of antibiotics, A. baumannii antibiotic-resistant strains rapidly transfer the resistance to the susceptible isolates and causing the occurrence of MDR (23). In 2017, SENTRY Antimicrobial Surveillance Program reported the highest number of MDR strains of A. baumannii in Europe and Latin America, Asia-Pacific, and North America between 1997 and 2016 (24). Also, high prevalence of MDR A. baumannii has emerged as a serious problem in healthcare settings in Iran (25). Previous studies have shown that fluoroquinolones are one of the first-line therapies for A. baumannii infections (26). But several studies revealed a considerable increase in ciprofloxacin resistance in Iran (27, 28).
In the present study, among 55 strains of A. baumannii isolated from different clinical samples of patients admitted in the ICU of Milad Hospital in Tehran, Iran, the highest antibiotic resistance was related to ciprofloxacin (100%) and then meropenem and imipenem. The results of this study also showed that 96.36% of A. baumannii isolates were resistant to 3 or more classes of antibiotics and mentioned as MDR isolates. Nowroozi et al. in 2014 reported that the resistance of A. baumannii strains to amikacin, ciprofloxacin, cotrimoxazole, ceftazidime, and ceftriaxone was equal to 100%, while their resistance to gentamicin and tetracycline was equal to 86.1%. In addition, 100% of isolates were recognized as MDR strains (29).
In 2016, Sarhaddi et al. investigated the drug resistance pattern of carbapenem-resistant A. baumannii strains isolated from the burning department of hospitals in northeastern Iran. They concluded that all isolates were resistant to β-lactam antibiotics and ciprofloxacin (30). Nourbakhsh et al. in 2018 demonstrated that the antibiotic resistance pattern for A. baumannii isolates from Burn Center of Isfahan Hospital showed high resistance to ciprofloxacin, ceftazidime, and tetracycline with a frequency of 82.5%, 75.3%, 72%, respectively (31). The review study of Hamzeh et al. on antibiotic-resistant clinical A. baumannii isolates from Iran during 2012 - 2017, demonstrated that there was a significant increase in resistance to many antibiotics such as gentamicin, imipenem, meropenem, piperacillin, ampicillin/sulbactam, ticarcillin, tobramycin, and aztreonam (32).
Also, in 2019, Sedaghat et al. showed that all A. baumannii isolates from burn patients in Northeast of Iran were MDR due to considerable resistance to fluoroquinolones (95%), cephalosporins (93% - 98%), penicillins (97%), carbapenems (94% - 95%), and beta-lactamase inhibitors (87% - 100%) (33). Both intrinsic and acquired mechanisms can cause resistance in Acinetobacter (34, 35). Resistance to quinolones can be caused by different ways; one of them is an alternation in the bacterial efflux pump expression. Determining ciprofloxacin resistance in A. baumannii isolates was performed both in the presence and in the absence of efflux pump inhibitors (36, 37). In this study, the effect of the efflux pump and its role in the development of the resistance to ciprofloxacin was investigated using the CCCP inhibitor. It was shown that among 55 ciprofloxacin-resistant strains, only 3 strains exhibited a 4-fold MIC reduction. These strains were reported as strains with a high phenotypic expression of the efflux pump.
Previous studies have proven the emergence of MDR by these pumps in Escherichia coli strains (38). Adabi et al., in a study conducted in Tehran in 2015, reported that 8% of Pseudomonas aeruginosa isolates indicated a 4-fold decrease in amikacin MIC in the presence of CCCP reflecting the role of the efflux pumps in the development of drug resistance in P. aeruginosa isolates (39). The study of Nikasa et al. in 2013, showed that a 16-fold reduction was observed in MIC of ciprofloxacin among A. baumannii isolates after using the CCCP efflux pump inhibitor (40). Ardebili et al. in 2014 showed that all strains of Acinetobacter are resistant to ciprofloxacin with MIC values ranging from 4 to 128 μg/mL or more. Moreover, the strains’ susceptibility to ciprofloxacin increased in the presence of CCCP efflux pump inhibitor such that a 2 - 64-fold decrease was observed in 86.1% of the strains and they mentioned efflux-based system may play a role in fluoroquinolone resistance in A. baumannii isolates (41).
In 2018, Abbasi Shaye demonstrated that among forty-six clinical Acinetobacter isolates collected from 2 teaching hospitals of Mashhad, Iran, 20 A. baumannii isolates showed a 2-fold or higher reduction in amikacin MIC in the presence of CCCP (42). In a study by Ardehali et al. in 2019, the results of phenotypic detection of efflux pumps using CCCP efflux pump inhibitor revealed that 23.07% of tigecycline-resistant A. baumannii isolates could contain active efflux pumps. The results of their study indicate that RND-type efflux pumps appear to play a significant role in the tigecycline resistance of A. baumannii (43).
Therefore, the results of our study showed that mechanisms other than the efflux pumps may be involved in the resistance to ciprofloxacin. Studies demonstrated that mutation in the QRDR region and gyrA and parC genes is another important mechanism related to the resistance to ciprofloxacin and associated with high resistance to fluoroquinolones (44). In this study, mutations in gyrA and parC genes were studied among some ciprofloxacin-resistant A. baumannii isolates. The presence of gyrA and parC genes was reported in all of the studied strains. In A. baumannii isolates, which were resistant to ciprofloxacin with MIC ≥ 4µg/mL, just a mutation in parC gene (84 position mutation L > S) was shown that altered amino acid. Also, one isolate of A. baumannii, which were resistant to ciprofloxacin with MIC ≥ 4µg/mL showed a mutation in gyrA gene, (345 position mutatiom T > C), but the mutation did not alter the amino acid. Our results are relatively similar to Wisplinghoff et al. study in 2003 that among 147 ciprofloxacin-resistant A. baumannii isolates sequenced for QRDR regions, no mutation leading to resistance was observed, and they suggested that other mechanisms may involve in resistance (45).
In Valentine et al. study, the sequencing results of ciprofloxacin-resistant A. baumannii strains showed gyrA gene mutation in all the resistant strains. They declared that this mutation probably causes fluoroquinolone resistance, such as levofloxacin (46). In Iran, Ardebili et al. in 2015 found that mutation in gyrA and parC genes could be effective in the A. baumannii resistance to ciprofloxacin. The nucleotide sequencing results revealed that 45 (90%) of 50 isolates had amino acid alteration gyrA and parC as follow: 1 (2.2%) isolate in gyrA, 2 (4.4%) in parC gene and 42 (93.3%) in gyrA and parC concurrently [20]. Warner et al. in 2016 reported the high levels of mutation in the gyrA and parC genes in ciprofloxacin-resistant A. baumannii strains (47). In 2014, Fazeli et al. identified gyrA gene in 70 strains of A. baumannii isolated from the patients admitted to the ICU of Alzahra Hospital in Isfahan, Iran (48). Khayat et al., in a study in 2017, showed that all the ciprofloxacin-resistant Acinetobacter strains had a mutation in gyrA gene, but no mutation was observed in the parC gene (49). In a study by Nowroozy et al. in 2014, all A. baumannii strains had MIC ≥ 32 μg/mL, but gyrA gene mutation was detected in MIC ≥ 4μg/mL and for parC was MIC ≥ 32 μg/mL (27, 29).
5.1. Conclusions
This study showed that there is a high prevalence of A. baumannii MDR strains; thus, we can conclude that resistance to ciprofloxacin is common in all clinical isolates of A. baumannii. So due to the crucial role of A. baumannii in nosocomial infections, particularly in ICUs, it is necessary to apply appropriate strategies to control the spread of bacterial resistance. Also, the results of the present study show the MIC reduction of ciprofloxacin among A. baumannii isolates in the presence of the efflux pump CCCP inhibitor and 3 isolates have a high phenotypic activity of efflux pump. In this study, there is no association between gyrA and parC gene mutation and ciprofloxacin resistance. Therefore, the role of mechanisms other than alterations in gyrA and parC to decrease the susceptibility to quinolones in A. baumannii isolates such as expression of genes encoding efflux pumps proteins should be considered. Further studies with a larger number of isolates are required to clarify the mechanisms associated with resistance of A. baumannii.
Acknowledgements
References
-
1.
Zhang J, Zhao C, Chen H, Li H, Wang Q, Wang Z, et al. A multicenter epidemiology study on the risk factors and clinical outcomes of nosocomial intra-abdominal infections in China: results from the Chinese Antimicrobial Resistance Surveillance of Nosocomial Infections (CARES) 2007-2016. Infect Drug Resist. 2018;11:2311-9. [PubMed ID: 30532564]. [PubMed Central ID: PMC6245374]. https://doi.org/10.2147/IDR.S182180.
-
2.
Tan TY, Poh K, Ng SY. Molecular typing of imipenem-resistant Acinetobacter baumannii-calcoaceticus complex in a Singapore hospital where carbapenem resistance is endemic. Infect Control Hosp Epidemiol. 2007;28(8):941-4. [PubMed ID: 17620241]. https://doi.org/10.1086/518964.
-
3.
Houang ET, Chu YW, Leung CM, Chu KY, Berlau J, Ng KC, et al. Epidemiology and infection control implications of Acinetobacter spp. in Hong Kong. J Clin Microbiol. 2001;39(1):228-34. [PubMed ID: 11136776]. [PubMed Central ID: PMC87707]. https://doi.org/10.1128/JCM.39.1.228-234.2001.
-
4.
Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, et al. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect. 2018;64(3):282-90. [PubMed ID: 30532564]. https://doi.org/10.1016/j.jinf.2011.12.008.
-
5.
Fishbain J, Peleg AY. Treatment of Acinetobacter infections. Clin Infect Dis. 2010;51(1):79-84. [PubMed ID: 20504234]. https://doi.org/10.1086/653120.
-
6.
Chusri S, Chongsuvivatwong V, Rivera JI, Silpapojakul K, Singkhamanan K, McNeil E, et al. Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob Agents Chemother. 2014;58(7):4172-9. [PubMed ID: 24820079]. [PubMed Central ID: PMC4068534]. https://doi.org/10.1128/AAC.02992-14.
-
7.
Doi Y, Murray GL, Peleg AY. Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin Respir Crit Care Med. 2015;36(1):85-98. [PubMed ID: 25643273]. [PubMed Central ID: PMC4465586]. https://doi.org/10.1055/s-0034-1398388.
-
8.
Urban C, Segal-Maurer S, Rahal JJ. Considerations in control and treatment of nosocomial infections due to multidrug-resistant Acinetobacter baumannii. Clin Infect Dis. 2003;36(10):1268-74. [PubMed ID: 12746772]. https://doi.org/10.1086/374847.
-
9.
Hamouda A, Amyes SG. Novel gyrA and parC point mutations in two strains of Acinetobacter baumannii resistant to ciprofloxacin. J Antimicrob Chemother. 2004;54(3):695-6. [PubMed ID: 15282231]. https://doi.org/10.1093/jac/dkh368.
-
10.
Gales AC, Jones RN, Forward KR, Linares J, Sader HS, Verhoef J. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY Antimicrobial Surveillance Program (1997-1999). Clin Infect Dis. 2001;32 Suppl 2:S104-13. [PubMed ID: 11320451]. https://doi.org/10.1086/320183.
-
11.
Fierobe L, Lucet JC, Decre D, Muller-Serieys C, Deleuze A, Joly-Guillou ML, et al. An outbreak of imipenem-resistant Acinetobacter baumannii in critically ill surgical patients. Infect Control Hosp Epidemiol. 2001;22(1):35-40. [PubMed ID: 11198020]. https://doi.org/10.1086/501822.
-
12.
Singh H, Thangaraj P, Chakrabarti A. Acinetobacter baumannii: A Brief Account of Mechanisms of Multidrug Resistance and Current and Future Therapeutic Management. J Clin Diagn Res. 2013;7(11):2602-5. [PubMed ID: 24392418]. [PubMed Central ID: PMC3879836]. https://doi.org/10.7860/JCDR/2013/6337.3626.
-
13.
Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42-51. [PubMed ID: 25435309]. https://doi.org/10.1038/nrmicro3380.
-
14.
Van Bambeke F, Balzi E, Tulkens PM. Antibiotic efflux pumps. Biochem Pharmacol. 2000;60(4):457-70. [PubMed ID: 10874120]. https://doi.org/10.1016/s0006-2952(00)00291-4.
-
15.
Ni W, Li Y, Guan J, Zhao J, Cui J, Wang R, et al. Effects of Efflux Pump Inhibitors on Colistin Resistance in Multidrug-Resistant Gram-Negative Bacteria. Antimicrob Agents Chemother. 2016;60(5):3215-8. [PubMed ID: 26953203]. [PubMed Central ID: PMC4862505]. https://doi.org/10.1128/AAC.00248-16.
-
16.
Al-Harmoosh RA, Jarallah EM, Al-Shamari AM, Al-Khafaji HM. Detection of Efflux Pumps Genes in Clinical Isolates of Acinetobacter baumannii. Res J Pharm Technol. 2017;10(12):4231. https://doi.org/10.5958/0974-360x.2017.00775.2.
-
17.
Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56(1):20-51. [PubMed ID: 15914491]. https://doi.org/10.1093/jac/dki171.
-
18.
Cambau E, Gutmann L. Mechanisms of resistance to quinolones. Drugs. 1993;45 Suppl 3:15-23. [PubMed ID: 7689446]. https://doi.org/10.2165/00003495-199300453-00005.
-
19.
Ling TK, Ying CM, Lee CC, Liu ZK. Comparison of antimicrobial resistance of Acinetobacter baumannii clinical isolates from Shanghai and Hong Kong. Med Princ Pract. 2005;14(5):338-41. [PubMed ID: 16103700]. https://doi.org/10.1159/000086932.
-
20.
Ardebili A, Lari AR, Beheshti M, Lari ER. Association between mutations in gyrA and parC genes of Acinetobacter baumannii clinical isolates and ciprofloxacin resistance. Iran J Basic Med Sci. 2015;18(6):623-6. [PubMed ID: 26221488]. [PubMed Central ID: PMC4509960].
-
21.
Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis. 2010;10:196. [PubMed ID: 20609238]. [PubMed Central ID: PMC2909240]. https://doi.org/10.1186/1471-2334-10-196.
-
22.
Karbasizade V, Heidari L. [Antimicrobial resistance of Acinetobacter baumannii isolated from Intensive Care Units of Isfahan hospitals, Iran]. J Isfahan Med School. 2012;30(191). Persian.
-
23.
Bhatt P, Rathi K, Hazra S, Sharma A, Shete V. Prevalence of multidrug resistant Pseudomonas aeruginosa infection in burn patients at a tertiary care centre. Indian J Burns. 2015;23(1):56. https://doi.org/10.4103/0971-653x.171656.
-
24.
SENTRY Program,. 2019. Available from: https://www.jmilabs.com/data/posters/ECCMID2018-SENTRY-Acinetobacter.pdf.
-
25.
Shirmohammadlou N, Zeighami H, Haghi F, Kashefieh M. Resistance pattern and distribution of carbapenemase and antiseptic resistance genes among multidrug-resistant Acinetobacter baumannii isolated from intensive care unit patients. J Med Microbiol. 2018;67(10):1467-73. [PubMed ID: 30175954]. https://doi.org/10.1099/jmm.0.000826.
-
26.
Prashanth K, Badrinath S. In vitro susceptibility pattern of Acinetobacter species to commonly used cephalosporins, quinolones, and aminoglycosides. Indian J Med Microbiol. 2004;22(2):97-103. [PubMed ID: 17642704].
-
27.
Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: A study in western Iran. Iran J Microbiol. 2013;5(3):195-202. [PubMed ID: 24475323]. [PubMed Central ID: PMC3895554].
-
28.
Rahbar M, Mehrgan H, Aliakbari NH. Prevalence of antibiotic-resistant Acinetobacter baumannii in a 1000-bed tertiary care hospital in Tehran, Iran. Indian J Pathol Microbiol. 2010;53(2):290-3. [PubMed ID: 20551535]. https://doi.org/10.4103/0377-4929.64333.
-
29.
Nowroozi J, Akhavan Sepahi A, Tahmasebinejad Kamarposhti L, Razavipour R, Mazhar F. Evaluation of Ciprofloxacin (gyrA, parC Genes) and Tetracycline (tetB Gene) Resistance in Nosocomial Acinetobacter baumannii Infections. Jundishapur J Microbiol. 2014;7(2). e8976. [PubMed ID: 25147676]. [PubMed Central ID: PMC4138684]. https://doi.org/10.5812/jjm.8976.
-
30.
Sarhaddi N, Dolatabadi S, Amel Jamehdar S. [Drug resistance pattern of carbapenem-resistant Acinetobacter baumannii isolated from a referral burn center in northeast of Iran]. Med J Mashhad. 2016;59:1-8. Persian.
-
31.
Nourbakhsh V, Borooni S, Daneshmand D, Tajbakhsh E, Nourbakhsh F. Antibiotic Resistance Patterns and Molecular Typing of Acinetobacter Baumannii Strains Isolated from Burn Patients in Iran. Int J Med Lab. 2018. https://doi.org/10.18502/ijml.v5i4.153.
-
32.
Hamzeh A, Rezaee P, Mohammadi M. Acinetobacter baumannii antibiotics resistance in Iran. J Bacteriol Mycol. 2019;7(6):159-62.
-
33.
Sedaghat A, Khadem-Rezaiyan M, Ahmadabadi A, Abbaspour H, Youssefi M, Shirzad MM, et al. Antibacterial Resistance Pattern of Acinetobacter baumannii in Burn Patients in Northeast of Iran. Jundishapur J Microbiol. 2019;12(10). https://doi.org/10.5812/jjm.94668.
-
34.
Szejbach A, Mikucka A, Bogiel T, Gospodarek E. Usefulness of phenotypic and genotypic methods for metallo-beta-lactamases detection in carbapenem-resistant Acinetobacter baumannii strains. Med Sci Monit Basic Res. 2013;19:32-6. [PubMed ID: 23333953]. [PubMed Central ID: PMC3638691]. https://doi.org/10.12659/msmbr.883744.
-
35.
Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568-85. [PubMed ID: 25857949]. https://doi.org/10.1016/j.ijantimicag.2015.03.001.
-
36.
Blanchard C, Barnett P, Perlmutter J, Dunman PM. Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob Agents Chemother. 2014;58(11):6360-70. [PubMed ID: 25114126]. [PubMed Central ID: PMC4249429]. https://doi.org/10.1128/AAC.03535-14.
-
37.
Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D'Souza S, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014;58(2):722-33. [PubMed ID: 24247144]. [PubMed Central ID: PMC3910843]. https://doi.org/10.1128/AAC.01866-13.
-
38.
Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45(5):1515-21. [PubMed ID: 11302820]. [PubMed Central ID: PMC90498]. https://doi.org/10.1128/AAC.45.5.1515-1521.2001.
-
39.
Adabi M, Talebi-Taher M, Arbabi L, Afshar M, Fathizadeh S, Minaeian S, et al. Spread of Efflux Pump Overexpressing-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas aeruginosa by using an Efflux Pump Inhibitor. Infect Chemother. 2015;47(2):98-104. [PubMed ID: 26157587]. [PubMed Central ID: PMC4495281]. https://doi.org/10.3947/ic.2015.47.2.98.
-
40.
Nikasa P, Abdi-Ali A, Rahmani-Badi A, Al-Hamad A. In vitro Evaluation of Proton Motive Force-Dependent Efflux Pumps Among Multidrug Resistant Acinetobacter baumannii Isolated From Patients at Tehran Hospitals. Jundishapur J Microbiol. 2013;6(7). https://doi.org/10.5812/jjm.6792.
-
41.
Ardebili A, Talebi M, Azimi L, Rastegar Lari A. Effect of Efflux Pump Inhibitor Carbonyl Cyanide 3-Chlorophenylhydrazone on the Minimum Inhibitory Concentration of Ciprofloxacin in Acinetobacter baumannii Clinical Isolates. Jundishapur J Microbiol. 2014;7(1). e8691. [PubMed ID: 25147654]. [PubMed Central ID: PMC4138672]. https://doi.org/10.5812/jjm.8691.
-
42.
Abbasi Shaye M, Sharifmoghadam MR, Bahreini M, Ghazvini K, Mafinezhad A, Amiri G. Study of the Role of Efflux Pumps in Amikacin-Resistant Acinetobacter Isolates from Teaching Hospitals of Mashhad, Iran. Jundishapur J Microbiol. 2018;11(4). https://doi.org/10.5812/jjm.12754.
-
43.
Ardehali SH, Azimi T, Fallah F, Owrang M, Aghamohammadi N, Azimi L. Role of efflux pumps in reduced susceptibility to tigecycline in Acinetobacter baumannii. New Microbes New Infect. 2019;30:100547. [PubMed ID: 31193724]. [PubMed Central ID: PMC6541740]. https://doi.org/10.1016/j.nmni.2019.100547.
-
44.
Hamouda A, Amyes SG. Development of highly ciprofloxacin-resistant laboratory mutants of Acinetobacter baumannii lacking topoisomerase IV gene mutations. J Antimicrob Chemother. 2006;57(1):155-6. [PubMed ID: 16287982]. https://doi.org/10.1093/jac/dki397.
-
45.
Wisplinghoff H, Decker M, Haefs C, Krut O, Plum G, Seifert H. Mutations in gyrA and parC associated with resistance to fluoroquinolones in epidemiologically defined clinical strains of Acinetobacter baumannii. J Antimicrob Chemother. 2003;51(1):177-80. [PubMed ID: 12493807]. https://doi.org/10.1093/jac/dkf254.
-
46.
Valentine SC, Contreras D, Tan S, Real LJ, Chu S, Xu HH. Phenotypic and molecular characterization of Acinetobacter baumannii clinical isolates from nosocomial outbreaks in Los Angeles County, California. J Clin Microbiol. 2008;46(8):2499-507. [PubMed ID: 18524965]. [PubMed Central ID: PMC2519477]. https://doi.org/10.1128/JCM.00367-08.
-
47.
Warner WA, Kuang SN, Hernandez R, Chong MC, Ewing PJ, Fleischer J, et al. Molecular characterization and antimicrobial susceptibility of Acinetobacter baumannii isolates obtained from two hospital outbreaks in Los Angeles County, California, USA. BMC Infect Dis. 2016;16:194. [PubMed ID: 27146090]. [PubMed Central ID: PMC4857389]. https://doi.org/10.1186/s12879-016-1526-y.
-
48.
Fazeli H, Vakili B, Khorvash F, Shoaei P, Kariminik A, Yaran M, et al. [Identification of mutation in gyrA gene obtained from quinolone-resistant clinical isolates of Acinetobacter baumannii]. J Microbial World. 2014;7(2):109-17. Persian.
-
49.
Khayat H, Sadeghifard N, Pakzad I, Azimi L, Delfani S, Sayehmiri K, et al. Determination of Different Fluoroquinolone Mechanisms Among Clinical Isolates of Acinetobacter baumannii in Tehran, Iran. Iran Red Crescent Med J. 2017;19(9). https://doi.org/10.5812/ircmj.58798.