Abstract
Background:
Anaerobic infections have been reported for many years, and there is an increasing trend in these infections worldwide, but anaerobic infections have not received sufficient attention. Rapid identification is important for the treatment of anaerobes because of their different antibiotic-resistance profiles.Objectives:
This study aimed to analyze the hospital’s present condition to improve anaerobic culture detection rates and enhance the monitoring of anaerobes in hospitals.Methods:
This study retrospectively analyzed sterile body fluids sent to the First Affiliated Hospital of Nanchang University in the form of culture bottles in 2017. Finally, 28 strains of obligate anaerobes were isolated, then combined 16S rRNA gene sequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry was used to identify the strains and performed separate antimicrobial susceptibility testing.Results:
The results showed that these two methods are highly consistent. There were 17/28 (61%) Gram-negative and 11/28 (39%) Gram-positive bacteria. The predominant bacteria were Bacteroides fragilis (15/28). Ten strains were isolated from the Obstetrics and Gynecology Department. The next most frequently affected departments were General Surgery (17.86%) and the ICU (17.86%). We analyzed the resistance to penicillin, cefoxitin, clindamycin, metronidazole, meropenem, piperacillin/tazobactam and amoxicillin/clavulanic acid by using the agar dilution method. The resistance rates to clindamycin were relatively high but relatively sensitive to metronidazole.Conclusions:
The results of this research indicate that we should pay attention to the cultivation of anaerobic bacteria, especially in certain high-risk departments.Keywords
Anaerobic Bacteria Identification Antimicrobial Susceptibility
1. Background
Anaerobic bacteria are one of the most important components of the normal flora and are widely present on human skin and mucosal surfaces, particularly in the intestinal tract and oral cavity. Anaerobic bacteria can cause a variety of life-threatening infections with high mortality rates (1) when the mucosal surface becomes disrupted and a plethora of anaerobic bacteria invade the deep tissue (2). In addition, factors that can reduce the oxygen concentration, such as aerobic/facultative bacteria multiplying rapidly, also have the potential to cause infection. To date, anaerobic infections have not received sufficient attention. From the blood culture bottles submitted for clinical examination in our hospital in 2017, the difference between the number of aerobic and anaerobic cultures was quite large, indicating that there might be some diagnostic omissions.
Anaerobes can cause a great variety of diseases, such as tetanus caused by Clostridium tetani, genitourinary tract infections caused by Bacteroides fragilis and gas gangrene caused by C. perfringens infection. A study confirmed that Clostridium spp. are associated with diabetic foot infection, Veillonella parvula has been found in head and neck infections, and B. fragilis has been linked to aerobic bacterial pathogens (3). Gram-positive, anaerobic cocci (GPAC) are the predominant pathogens in human infections according to numerous studies. Species of GPAC can cause a severe infection, bacteremia. Parvimonas spp. are most frequently isolated from bacteremia specimens (4).
There are many reports on anaerobic infections worldwide, and it has been demonstrated that antibiotics in the current market have reduced the efficacy against anaerobic bacterial infection (5). Antimicrobial resistance among anaerobes is increasing dramatically due to changes in the drug resistance mechanisms (6). Metronidazole is a broad-spectrum antibiotic for anaerobic infections. Gram-negative bacilli, such as B. fragilis, are susceptible to metronidazole. Nevertheless, increasing resistance rates in Propionibacterium and Actinomyces have been observed (7). Bacteroides and Prevotella species have a high level of resistance to penicillin and ampicillin (7). Resistance to clindamycin is increasing obviously around the world. Imipenem is active against the overwhelming majority of anaerobes. Therefore, antimicrobial susceptibility testing plays an important role in antibacterial therapy because the empirical medication may not have satisfactory effects in clinical practice.
Compared with aerobic bacteria, anaerobic bacteria are significantly more difficult to cultivate and identify (8). Several methods have been applied for the identification of anaerobic bacteria. Conventional methodology is time-consuming with low specificity (9). To meet the increasing requirement for the identification of anaerobes, new automated identification methods have been proposed in recent years. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) is one such method. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry can shorten the identification time and ensure identification with a certain accuracy level, but it is still limited. More data are needed about clinical isolates to verify these new methods before they can be applied to routine microorganism identification (10), and the mass spectral database of bacteria is limited. Moreover, 16S rRNA gene sequencing is increasingly used in microbial identification in clinical microbiology laboratories. This method can identify bacteria to the species level with a relatively high level of accuracy. However, it has a disadvantage in terms of time (11).
2. Objectives
We combined the characteristics of these methods in our study. We isolated anaerobic bacteria from the First Affiliated Hospital of Nanchang University in 2017 and identified the bacteria with a combination of MALDI-TOF MS and 16S rRNA gene sequencing, analyzing the distribution characteristics between different departments and drug resistance in this hospital. We aimed to conduct a retrospective study to provide clinical treatment advice.
3. Methods
3.1. Patients and Isolates
We collected 30,469 isolates from the First Affiliated Hospital of Nanchang University in 2017. The samples included blood, cerebrospinal fluid, pleural fluid, ascites, and other sterile body fluids, which were linked with patient data gathered from the electronic medical record. The samples were collected by clinicians or nurses and then transferred to the clinical microbiology laboratory. Aerobic, anaerobic or fungal culturing bottles were performed, and the duplicate samples were removed. When blood culture instrument alarm, the positive bottles were removed and then the specimens were extracted using a syringe, inoculated in blood agar plates and chocolate agar plates, incubated at 35ºC in anaerobic environment using a rapid and automatic anaerobic cultivation system (Anoxomat MART Microbiology B.V., The Netherlands).
3.2. Identification of Anaerobes
In this study, 16S rRNA gene sequencing and MALDI-TOF MS (Bruker, Sancordon Inc., Bremen, Germany) were used to identify the strains. The 16S rRNA gene was amplified by PCR after extraction of the bacterial genome. The primers of the PCR are shown in Table 1, and the PCR was performed in a final volume of 25 μL containing 12.5 μL of 2xTaq PCR Mastermix, 1 µL primer 1 (10 µmol/L), 1 µL of primer 2 (10 µmol/L), 1 µL of template DNA (20 ng/μL), and add pure water to 25µL. The reaction conditions were as follows: 95ºC for 5min, followed by 35 cycles of 95ºC for 30 s, 56ºC for 40 s, and 72ºC for 2 min, and finally, 72ºC for 5min and stored at 4ºC. All the 16 s rRNA PCR products were electrophoresed on 1.5% agarose gel with ethidium bromide staining (100 V, 40 min) and sequenced using high-throughput sequencing technology.
The sequences were then analyzed. A ≥ 99% match with the 16S rRNA gene sequence was confirmed for species-level identification. The MALDI-TOF MS processes were as follows: A single bacterial colony that had been cultured for 24 - 48 h was transferred to the MALDI target plates by using a sterile toothpick. Then, 0.5 µL of 70% formic acid was added to the colony, 1 µL of 4-hydroxycinnamic acid (HCCA) matrix solution was used to overlay the microbial film, allowed to dry at room temperature and finally analyzed with MALDI-TOF MS. A log score of ≥ 1.7 was considered indicative of high confidence.
Information About the 16S rRNA Primers
| Gene Primer | Sequence (5’ - 3’) | Length |
|---|---|---|
| 16S rRNA 27f | AGAGTTTGATCCTGGCTCAG | 1510 bp |
| 1492r | CTACGGCTACCTTGTTACGA |
3.3. Antimicrobial Susceptibility Testing
Microbial susceptibility to penicillin, cefoxitin, clindamycin, metronidazole, meropenem, piperacillin/tazobactam and amoxicillin/clavulanic acid was determined by using the agar dilution method. In this method, Brucella agar supplemented with 5% dissolved sheep blood, 5 µg/mL hemin and 1 µg/mL Vitamin K1 was used. Antibiotics at different concentrations were added to 50ºC Brucella agar (that had been melted), which was then poured into plates and allowed to solidify. A standardized number of bacteria were inoculated onto the plates using inoculators. At 30 minutes after inoculation, the plates were placed in an anaerobic incubator with 5% CO2 at 35ºC to culture for 48 - 72 h. The minimal inhibitory concentration (MIC) was defined as the lowest concentration of the antibiotic that repressed visible growth. The results were read according to the CLSI M100 guidelines (2017). In the course of the experiment, we conducted quality controls using B. fragilis ATCC25285, and the β-lactamase assay was performed using cephalosporin sheets.
4. Results
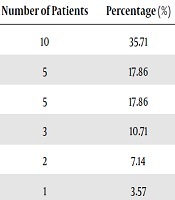
In 2017, a total of 30,469 clinical isolates were collected. There were 10,406 anaerobic culture bottles, 18,094 aerobic culture bottles and 1,969 fungal culture bottles. After culturing, 831 samples from 29 departments showed positive cultures in anaerobic culture bottles, and 803 of these strains were aerobic or facultative bacteria. Finally, a total of 28 strains were anaerobic bacteria. The positivity rate of obligate anaerobic culture was 3.37% (28/831). There were 17/28 (61%) Gram-negative and 11/28 (39%) Gram-positive bacteria. In our research, the predominant anaerobic species isolated was B. fragilis, accounting for nearly half of those identified. The majority of the samples were blood. Other bacteria isolated include C. difficile (7.14%), Anaerococcus tetradius (7.14%), and Slackia exigua (3.57%). Also, 28 strains of anaerobic bacteria were isolated from eight departments. The specific information is listed in Table 2.
The Distribution of Samples and Anaerobes in Different Departments
| Department | Characteristic | Samples | ||||
|---|---|---|---|---|---|---|
| Number of Patients | Percentage (%) | bl | ab | sf | Others | |
| Obstetrics and gynecology | 10 | 35.71 | 10 | |||
| General surgery | 5 | 17.86 | 4 | 1 | ||
| ICU | 5 | 17.86 | 5 | |||
| EICU | 3 | 10.71 | 3 | |||
| Gastroenterology | 2 | 7.14 | 2 | |||
| Neurology | 1 | 3.57 | 1 | |||
| Respiratory | 1 | 3.57 | 1 | |||
| Orthopedics | 1 | 3.57 | 1 | |||
| Total | 28 | 100 | 23 | 3 | 1 | 1 |
Obstetrics and gynecology was the dominant department, and general surgery and ICU were the second most common departments with anaerobic bacteria. In addition to this, obstetrics and gynecology, ICU, EICU and respiratory, the source of the four departments was blood. The general surgery department collected five strains, four of them were from blood, and the other one were collected from ascites. Besides, there were two ascite samples from gastroenterology, one cerebrospinal fluid sample from neurology department and one other sterile body fluids specimen from the orthopaedics department. MALDI-TOF MS provided species-level identification for 27 (27/28) isolates, with only one strain not being identified. However, the identification results of these 27 strains were completely consistent with the 16S rRNA gene sequencing results. The results of the antimicrobial susceptibility testing are shown in Table 3. Clindamycin resistance was the most common among the tested antibiotics, with 18 strains resistant to it. In contrast, metronidazole was the most effective antibiotic in this research. In addition, the B. fragilis and C. difficile isolates were resistant to both cefoxitin and clindamycin.
The Results of Antimicrobial Susceptibility Testinga
| Anaerobic bacteria Total | Clindamycin, R(n) | Metronidazole, R(n) | Penicillin, R(n) | Cefoxitin, R(n) | Meropenem, R(n) | Piperacillin/Tazobactam, R(n) | Amoxicillin/Clavulanic Acid, R(n) | |
|---|---|---|---|---|---|---|---|---|
| Gram negative bacteria | ||||||||
| Bacteroides fragilis | 15 | 12 | 0 | N/A | 5 | 5 | 5 | 9 |
| Bacteriodes vulgatus | 1 | 1 | 0 | 1 | N/A | N/A | N/A | N/A |
| Prevotella amnii | 1 | 0 | 0 | 0 | N/A | N/A | N/A | N/A |
| Gram positive bacteria | ||||||||
| Cutibacterium acnes | 2 | 0 | 2 | 0 | N/A | N/A | N/A | N/A |
| Anaerococcus tetradius | 2 | 1 | 0 | 0 | N/A | N/A | N/A | N/A |
| Staphylococcus saccharolyticus | 1 | 0 | 0 | 0 | N/A | N/A | N/A | N/A |
| Anaerococcus hydrogenalis | 1 | 0 | 0 | 0 | N/A | N/A | N/A | N/A |
| Slackia exigua | 1 | 0 | 0 | 0 | N/A | N/A | N/A | N/A |
| Clostridium difficile | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 0 |
| Clostridium innocuum | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 |
| Clostridium saccharolyticum | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Drug resistance rate (r%) | 64% | 7% | 31% | 42% | 26% | 26% | 47% |
5. Discussion
The rising global trend in anaerobic bacterial infection has drawn attention. Accurate identification is essential for the treatment because anaerobes are sensitive to many antibiotics. In our study, we collected 30,469 samples from the clinic in 2017. The samples were much less than Rassolie and Ozenci (12) collected from Klinisk Mikrobiologi in Huddinge University Hospital. There were 18,094 isolates submitted for aerobic culture; however, only 10,406 isolates were submitted for anaerobic culture. Therefore, aerobic and anaerobic cultures are not performed at a one-to-one ratio in our hospital. This reduced rate of anaerobic cultures can increase the number of undetected anaerobes and make the treatment of bacterial infections difficult. However, using our statistical analysis and the presentations of clinical departments, this situation improved in the first half of 2018, and the gap between anaerobic and aerobic referral rates is narrowing.
The blood samples (26,740/30,469) were the most abundant, and anaerobic bacteria grew in 0.09% (23/26740) of these samples. The prevalence of anaerobic bacteria in these samples is higher than that in the research conducted by Gross et al. (13) who analyzed blood cultures taken in the pediatric emergency department from 2002 to 2016, and found obligate anaerobic bacteria in 33 cultures (0.05%). However, in a three-year study, Rassolie and Ozenci collected blood culture bottles at a tertiary care hospital at a rate of 0.76% (12). Bacteroides fragilis was the predominant isolate in our study, mirroring the results of other studies (3, 14). The next most frequently isolated bacteria were C. difficile, A. tetradius, Cutibacterium acnes and others, which account for less than half of the total, which is different from previous reports (12).
The identification of bacterial species by MALDI-TOF MS was highly consistent with that of the 16S rRNA gene sequencing in our study, and only one strain was not identified. The unidentified strain was A. hydrogenalis. Previous studies show that A. hydrogenalis is predominantly identified by gene sequencing (15-17), and there have been only a few reports about A. hydrogenalis isolated from vaginal discharge and ovarian abscesses (17). In this study, we prefer gene sequencing to MALDI-TOF MS for A. hydrogenalis identification. Several reasons for this preference are related to the limitations of MALDI-TOF MS. First, different stages of bacterial growth can present different protein expression levels, which may affect detectability since MALDI-TOF MS analyzes proteins (18). Second, insufficient reference spectra can lead to misleading identifications, which is particularly common for anaerobes (19).
Seven antimicrobial agents were used in for antimicrobial susceptibility testing and showed different levels of resistance. The most frequent isolate in our research, strains of B. fragilis, were resistant to cefoxitin, clindamycin, meropenem, piperacillin/tazobactam and amoxicillin/clavulanic acid, with the exception of metronidazole, the susceptibility to which was similar to that of other studies (14). Multidrug-resistant (MDR) bacteria are a difficult problem worldwide. Multidrug resistance increases the difficulty of treating bacterial infections. Multidrug-resistant B. fragilis has been reported in many regions (20, 21). In our study, a total of five multidrug-resistant B. fragilis strains were isolated. Two strains were from the EICU, obstetrics and gynecology, ICU and general surgery, and each department had one strain. The prevalence of MDR B. fragilis is considered to be associated with resistance genes and the abuse of antibiotics (22).
In our study, only one Prevotella sp., P. amnii, was susceptible to clindamycin, but Prevotella spp. have an alarming resistance to clindamycin (23). Multidrug resistance should be considered when physicians choose agents to treat anaerobic infections. We also found that S. saccharolyticus, A. hydrogenalis and S. exigua were completely susceptible to clindamycin, metronidazole and penicillin in our research. Clindamycin was the least effective antibiotic, with 18/28 strains, including 6 species, being resistant. This result is in accordance with other studies (23). Bacteroides fragilis accounted for 67% (12/18) of the drug-resistant strains, and we isolated 15 B. fragilis strains in total. Therefore, the clindamycin-resistance rate in B. fragilis is 80%, which is higher than has been observed in other studies (14, 24). Metronidazole was the most effective antibiotic, and only two C. acnes strains were resistant.
The distribution of anaerobes in hospitals varies. In our study, the main departments affected were obstetrics and gynecology (35.71%), general surgery (17.86%) and the ICU (17.86%). As our data show, we collected 10 isolates from obstetrics and gynecology, which were all from blood samples. Why anaerobes were mainly collected from obstetrics and gynecology may be explained by the following reasons. The vagina and urogenital tract are colonized with a plethora of anaerobes, which are part of the normal flora together with aerobes and facultative anaerobes. Patients in the obstetrics and gynecology department sometimes have colposcopies and surgery. It is possible that the colposcopy can result in the transference of anaerobes from the urogenital tract to the bloodstream. The use of postoperative antibacterial agents also increases the risk of anaerobic infection. Bacterial vaginosis (BV) is the most common form of vaginal infection in women. The BV is often characterized by an overgrowth of Gardnerella vaginalis and other anaerobic bacteria, such as Atopobium vaginae, Bacteroides spp., Mobiluncus spp. and Prevotella spp. (25).
The infection rate of Trichomonas vaginitis has declined in recent years and has been replaced by pathogenic infections, which may be another reason why the majority of anaerobic bacteria were isolated from obstetrics and gynecology. Therefore, it was necessary to remind practitioners in the obstetrics and gynecology department to preoperatively use anti-anaerobic drugs. After the obstetrics and gynecology department applied our advice, metronidazole prophylaxis was used before some surgeries or colposcopies, and bloodstream infections by anaerobic bacteria were indeed significantly reduced. The frequent identification of anaerobic infections in the general surgery department is because of intestinal surgery, which facilitates anaerobic bacterial infections. Both the ICU and the EICU care for seriously ill patients, and patients with severe diseases are more likely to be infected with anaerobic bacteria because of their weakened immunity. We found that there was a significant difference in the anaerobes isolated from the various departments. Patients suffer from different diseases in each department. It stands to reason that pathogenesis varies according to individual cases; the types of sample also vary from person to person. These and other factors cause the difference. If a large amount of data about the characteristics of bacterial distribution in hospitals can be gained, clinicians can make empirical judgments according to the symptoms and examination.
5.1. Conclusions
The identification of anaerobic bacteria can be time-consuming because of their strict environmental growth requirements. However, anaerobes can cause many kinds of severe infections and have received much attention. We must improve anaerobic culture detection rates and enhance the monitoring of anaerobes in hospitals.
Acknowledgements
References
-
1.
Jeverica S, Nagy E, Mueller-Premru M, Papst L. Sample preparation method influences direct identification of anaerobic bacteria from positive blood culture bottles using MALDI-TOF MS. Anaerobe. 2018;54:231-5. [PubMed ID: 29861277]. https://doi.org/10.1016/j.anaerobe.2018.05.003.
-
2.
Noor A, Khetarpal S. Anaerobic infections. StatPearls [Internet]. StatPearls Publishing; 2019.
-
3.
Shenoy PA, Vishwanath S, Gawda A, Shetty S, Anegundi R, Varma M, et al. Anaerobic bacteria in clinical specimens - frequent, but a neglected lot: A five year experience at a tertiary care hospital. J Clin Diagn Res. 2017;11(7):DC44-8. [PubMed ID: 28892897]. [PubMed Central ID: PMC5583845]. https://doi.org/10.7860/JCDR/2017/26009.10311.
-
4.
Badri M, Nilson B, Ragnarsson S, Senneby E, Rasmussen M. Clinical and microbiological features of bacteraemia with Gram-positive anaerobic cocci: A population-based retrospective study. Clin Microbiol Infect. 2019;25(6):760 e1-6. [PubMed ID: 30217761]. https://doi.org/10.1016/j.cmi.2018.09.001.
-
5.
Thakare R, Dasgupta A, Chopra S. Eravacycline for the treatment of patients with bacterial infections. Drugs Today (Barc). 2018;54(4):245-54. [PubMed ID: 29869646]. https://doi.org/10.1358/dot.2018.54.4.2800623.
-
6.
Nagy E, Boyanova L, Justesen US; Escmid Study Group of Anaerobic Infections. How to isolate, identify and determine antimicrobial susceptibility of anaerobic bacteria in routine laboratories. Clin Microbiol Infect. 2018;24(11):1139-48. [PubMed ID: 29458156]. https://doi.org/10.1016/j.cmi.2018.02.008.
-
7.
Schuetz AN. Antimicrobial resistance and susceptibility testing of anaerobic bacteria. Clin Infect Dis. 2014;59(5):698-705. [PubMed ID: 24867792]. https://doi.org/10.1093/cid/ciu395.
-
8.
Gajdacs M, Spengler G, Urban E. Identification and antimicrobial susceptibility testing of anaerobic bacteria: Rubik's cube of clinical microbiology? Antibiotics (Basel). 2017;6(4). [PubMed ID: 29112122]. [PubMed Central ID: PMC5745468]. https://doi.org/10.3390/antibiotics6040025.
-
9.
Koba M, Szostek A, Konopa J. Limitation of usage of PicoGreen dye in quantitative assays of double-stranded DNA in the presence of intercalating compounds. Acta Biochim Pol. 2007;54(4):883-6. [PubMed ID: 18066405].
-
10.
Yunoki T, Matsumura Y, Yamamoto M, Tanaka M, Hamano K, Nakano S, et al. Genetic identification and antimicrobial susceptibility of clinically isolated anaerobic bacteria: A prospective multicenter surveillance study in Japan. Anaerobe. 2017;48:215-23. [PubMed ID: 28935196]. https://doi.org/10.1016/j.anaerobe.2017.09.003.
-
11.
Duskova M, Sedo O, Ksicova K, Zdrahal Z, Karpiskova R. Identification of lactobacilli isolated from food by genotypic methods and MALDI-TOF MS. Int J Food Microbiol. 2012;159(2):107-14. [PubMed ID: 23072695]. https://doi.org/10.1016/j.ijfoodmicro.2012.07.029.
-
12.
Rassolie A, Ozenci V. Short-term culture for rapid identification of anaerobic bacteria from blood cultures. Anaerobe. 2019;57:59-62. [PubMed ID: 30926438]. https://doi.org/10.1016/j.anaerobe.2019.03.014.
-
13.
Gross I, Gordon O, Abu Ahmad W, Benenson S, Piatkowski BS, Eventov-Friedman S, et al. Yield of anaerobic blood cultures in pediatric emergency department patients. Pediatr Infect Dis J. 2018;37(4):281-6. [PubMed ID: 28858042]. https://doi.org/10.1097/INF.0000000000001751.
-
14.
Jeverica S, Kolenc U, Mueller-Premru M, Papst L. Evaluation of the routine antimicrobial susceptibility testing results of clinically significant anaerobic bacteria in a Slovenian tertiary-care hospital in 2015. Anaerobe. 2017;47:64-9. [PubMed ID: 28433670]. https://doi.org/10.1016/j.anaerobe.2017.04.007.
-
15.
Song Y, Liu C, McTeague M, Vu A, Liu JY, Finegold SM. Rapid identification of Gram-positive anaerobic coccal species originally classified in the genus Peptostreptococcus by multiplex PCR assays using genus- and species-specific primers. Microbiology. 2003;149(Pt 7):1719-27. [PubMed ID: 12855723]. https://doi.org/10.1099/mic.0.26227-0.
-
16.
Hill KE, Davies CE, Wilson MJ, Stephens P, Lewis MA, Hall V, et al. Heterogeneity within the gram-positive anaerobic cocci demonstrated by analysis of 16S-23S intergenic ribosomal RNA polymorphisms. J Med Microbiol. 2002;51(11):949-57. [PubMed ID: 12448679]. https://doi.org/10.1099/0022-1317-51-11-949.
-
17.
Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51(Pt 4):1521-8. [PubMed ID: 11491354]. https://doi.org/10.1099/00207713-51-4-1521.
-
18.
Schrottner P, Gunzer F, Schuppel J, Rudolph WW. Identification of rare bacterial pathogens by 16S rRNA gene sequencing and MALDI-TOF MS. J Vis Exp. 2016;(113). [PubMed ID: 27500532]. [PubMed Central ID: PMC4993432]. https://doi.org/10.3791/53176.
-
19.
Croxatto A, Prod'hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36(2):380-407. [PubMed ID: 22092265]. https://doi.org/10.1111/j.1574-6976.2011.00298.x.
-
20.
Salipante SJ, Kalapila A, Pottinger PS, Hoogestraat DR, Cummings L, Duchin JS, et al. Characterization of a multidrug-resistant, novel Bacteroides genomospecies. Emerg Infect Dis. 2015;21(1):95-8. [PubMed ID: 25529016]. [PubMed Central ID: PMC4285247]. https://doi.org/10.3201/eid2101.140662.
-
21.
Ank N, Sydenham TV, Iversen LH, Justesen US, Wang M. Characterisation of a multidrug-resistant Bacteroides fragilis isolate recovered from blood of a patient in Denmark using whole-genome sequencing. Int J Antimicrob Agents. 2015;46(1):117-20. [PubMed ID: 25940770]. https://doi.org/10.1016/j.ijantimicag.2015.02.024.
-
22.
Sarvari KP, Soki J, Kristof K, Juhasz E, Miszti C, Melegh SZ, et al. Molecular characterisation of multidrug-resistant Bacteroides isolates from Hungarian clinical samples. J Glob Antimicrob Resist. 2018;13:65-9. [PubMed ID: 29101081]. https://doi.org/10.1016/j.jgar.2017.10.020.
-
23.
Xie Y, Chen J, He J, Miao X, Xu M, Wu X, et al. Antimicrobial resistance and prevalence of resistance genes of obligate anaerobes isolated from periodontal abscesses. J Periodontol. 2014;85(2):327-34. [PubMed ID: 23659425]. https://doi.org/10.1902/jop.2013.130081.
-
24.
Marchand-Austin A, Rawte P, Toye B, Jamieson FB, Farrell DJ, Patel SN. Antimicrobial susceptibility of clinical isolates of anaerobic bacteria in Ontario, 2010-2011. Anaerobe. 2014;28:120-5. [PubMed ID: 24923267]. https://doi.org/10.1016/j.anaerobe.2014.05.015.
-
25.
Jung HS, Ehlers MM, Lombaard H, Redelinghuys MJ, Kock MM. Etiology of bacterial vaginosis and polymicrobial biofilm formation. Crit Rev Microbiol. 2017;43(6):651-67. [PubMed ID: 28358585]. https://doi.org/10.1080/1040841X.2017.1291579.