Abstract
Background:
Atypical pneumonia is an upper and lower respiratory tract infection. Mycoplasma pneumoniae is a major cause of community-acquired pneumonia (CAP).Objectives:
The present study aimed to determine the prevalence of atypical pneumonia caused by M. pneumonia by culture and molecular PCR methods in Tehran.Methods:
In the present study, 102 samples of throat swab were collected from patients with respiratory infections. All samples were cultured in liquid PPLO Broth And solid PPLO agar media (1% glucose and 20% horse serum). The PCR technique with specific primers was implemented after culture and genome extraction through phenol-chloroform technique.Results:
In this study, 27 (26.47%) colonies of Mycoplasma were isolated on PPLO agar medium. Using specific primers, it was found that 33 samples (32.4%) were positive in terms of Mycoplasma genus and 14 samples (13.7%) were positive for the presence of M. pneumonia.Conclusions:
Mycoplasma pneumonia is a pathogen that causes respiratory tract infections in humans. Molecular PCR method is a quick and sensitive technique that has higher sensitivity and specificity than other methods. The obtained results may contribute to the specific treatment of some patients with symptoms of respiratory infections.Keywords
Atypical Pneumonia Mycoplasma pneumonia P1 Gene Culture and Molecular Methods
1. Background
Pneumonia is a lung parenchyma infection that is prevalent in any age group and is known as the sixth cause of death and the most common death-related infectious syndrome (1, 2). Atypical Pneumonia syndrome causes upper and lower respiratory tract infection that can be characterized by gradual onset, dry cough, shortness of breath, extraterrestrial symptoms (such as headache, muscle aches, fatigue, sore throat, nausea, vomiting, and diarrhea) (3-5). Mycoplasma pneumoniae is a major cause of community-acquired pneumonia. Mycoplasma pneumoniae is a parasite bacterium belonging to the Mollicutes class that has no cell wall and usually has small genomes less than 1000 kb (6-8). Mycoplasma pneumoniae bacterium encodes 116KD protein on its surface antigen that determines the serum activity of patients (9).
In M. pneumoniae, lipoproteins act as inflammatory agents. Recent reports suggest that M. pneumoniae also causes inflammatory responses independent of TLR2, TLR4, and autophagy (10). This common pathogen is common in the upper and lower respiratory tract of humans around the world and causes atypical pneumonia and tracheobronchitis (11, 12). Mycoplasma pneumoniae is known as the cause of atypical pneumonia, especially in children. This microorganism is reported in 5 to 40 percent of all acquired pneumonia in children aged 5 to 15 (13-16). Transmission of infection occurs through close contact with respiratory droplets. The clinical course of M. pneumoniae infections is usually mild and self-limiting and can be resolved without any treatment within 2 to 4 weeks (17-19). The annual incidence and prevalence of community-acquired pneumonia vary from 1.5 to 1.7 per 1000 adults in Europe. The community-acquired pneumonia is different from a mild or severe outpatient condition under that patients need to be admitted, and approximately 10% of patients need to be admitted to the intensive care unit (ICU).
In community-acquired pneumonia by M. pneumonia, up to 37% of patients receive ambulatory care and 10% of them need hospitalization (18). In a study on 585 cases of severe acute respiratory infection (SARI) and acute severe respiratory infection, the prevalence of pneumonia by M. pneumoniae was 19.66% and 58.15%. A study in the United States in the 1960s indicated that 50% of pneumonia in children and 35% in adults were due to both viral cause and M. pneumoniae, and a study in Japan in the 1980s indicated that 26.8% of children with pneumonia were positive in terms of the presence of M. pneumoniae [14]. Data of 21 countries indicated that M. pneumoniae was the most common type of bacterium responsible for atypical pneumonia that caused about 12% of community-acquired pneumonia during 1996 - 2004 (20). In a study by M. Maheshwari et al., it was determined that 75 patients suffered from an infection of a lower respiratory tract using PCR method. It was found that the prevalence of pneumonia due to M. pneumoniae was 30.7% (21). In a study on Iraq, the prevalence of pneumonia infection due to M. pneumoniae was estimated at 19.4% (22).
2. Objectives
Considering that a few studies have been conducted on this field based on molecular methods in Iran and most studies are based on culture and serological methods, further studies on methods with high sensitivity and precision such as PCR are very beneficial and important to present an accurate rate of the disease prevalence. Therefore, the present study aimed to determine the prevalence of atypical pneumonia caused by M. pneumoniae in Tehran by culture and molecular PCR methods.
3. Methods
3.1. Sample Collection
In the present descriptive-analytical study, 102 patients referred to Mostafa Khomeini and Khatam hospitals in Tehran were considered as the statistical population. They were approved and included in the study after the initial examination.
3.2. Bacterial Identification
3.2.1. Collection and Culture of Samples
A total of 102 samples of throat swab were collected from patients suspected with M. pneumoniae infections, including all patients with clinical symptoms of respiratory infection such as weakness and lethargy, fatigue, persistent headache and dry cough, shortness of breath, diarrhea, sputum, muscle pain, as well as removal of those, who consumed antibiotics, under the supervision of a pulmonologist according to sterility principles and conditions. After the samples in the transport medium were transferred to the laboratory, 1 mL of transport media was transferred, by passing through the 0.45 filter, to the main medium of PPLO broth (pH = 7.8 ± 0.2) and was incubated under CO2 5 - 10% for 3 weeks at 37 ℃. After 3 times of sub-culturing the samples in the liquid medium, 100 μL of samples was cultured in a solid PPLO agar medium (containing 1% Glucose and 20% horse serum) and the samples were incubated under CO2 5 - 10% at 37°C for 7 - 10 days (23). In the present study, M. pneumoniae (ATCC: 29342) was a standard strain that was obtained from Mycoplasma Reference Laboratory of Razi Vaccine and Serum Research Institute.
3.3. DNA Extraction
After cultivating clinical samples in the liquid medium of PPLO broth, the Phenol- chloroform method was used to extract DNA from liquid PPLO broth media in which the clinical samples were cultured. They were kept at -20°C after extracting DNA of samples until PCR.
3.4. Detection of Mycoplasma pneumoniae P1 Gene
In the present study, specific primers were used to identify the genus (16 Sr RNA gene) of Mycoplasma and species (P1 M. pneumoniae protein-encoding gene) (Table 1). After confirming the sensitivity and specificity of the primers by NCBI BLAST, the PCR reaction to the final volume of 25 was according to the protocol, and the following procedure was done: a primary denaturation cycle of 95°C for 5 minutes, 40 cycles including denaturation 95°C for 1 minute, a primer connection of 56°C for 1 minute, elongation of 72°C for 1 minute, and a final extension 72°C cycle for 5 minutes. Finally, the products of PCR reaction on 1% Agarose gel were compared to (standard strain M. pneumoniae strain ATCC: 29342) electrophoresis; and the accuracy of PCR method was approved.
Primer Sequences Used in This Study
| Gene Name | Primer Sequences | Produce Size (bp) | Resource |
|---|---|---|---|
| 16SrRNA | F: 5'- GCT GCG GTG AAT ACG TTC T -3' | 163 | The present study |
| R: 5'- TCCCCACGTTCTCGTAGGG -3' | |||
| P1 | F: 5'-AAAGGAAGCTGACTCCGACA-3' | 450 | (23) |
| R: 5'-TGGCCTTGCGCTACTAAGTT-3' |
4. Results
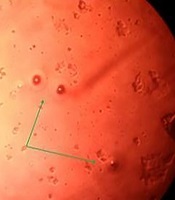
In the present study, 102 suspected cases with M. pneumoniae with a minimum age of 6, maximum age of 97 and an average age of 49 of whom 61 individuals were female (59.8%) and 41 were male (40.2%) and were tested by culture and molecular PCR methods. Of 102 samples, 27 samples (26.47%) were examined after four times of sub-cultivation in a culture medium, and then in a PPLO agar medium creating oocyte single colonies (Figure 1).
Mycoplasma pneumoniae colony

4.1. Results of PCR Launch for the Detection of Mycoplasma pneumoniae Genus and Species
The Mycoplasma genus (Mycoplasma 16SrRNA gene) was confirmed (Figure 2) in 33 cases (32.4%). Of the patients, 14 persons (13.7%) were infected with M. pneumoniae (Figure 3). In the present study, the molecular PCR method diagnosed 16S rRNA gene related to Mycoplasma genus (32.4%) and P1 gene relating to M. pneumoniae (13.7%) rapidly and accurately in patients with respiratory infections. In the culture on the PPLO agar medium (containing 1% Glucose and 20% horse serum), 27 cases (26.47%) were positive. The results indicated that the molecular PCR method had higher sensitivity and specificity. Also, sensitivity (66.67%) and specificity (94.74%) were reported for the molecular-PCR technique based on the analysis performed by the online statistical system medCalc 81. Most patients had generalized clinical symptoms including dry cough (19.6%), sputum cough (40.2%), fatigue (71.56%), lethargy (79.41%), headache (62.68%), nausea (20.58%), vomit (14.7%), and gastrointestinal involvement (36.27%) (Figure 4).
Gel electrophoresis of polymerase chain reaction amplification products for Mycoplasma 16SrRNA gene of 163 bp. Lane M, ladder (100 bp ladder); lane C+, positive control (ATCC :29342); lane C-, negative control; lane 1 - 6, positive samples.

Gel electrophoresis of polymerase chain reaction amplification products for Mycoplasma P1 gene of 450 bp. Lane M, ladder (100 bp ladder); lane C+, positive control (ATCC:29342), lane C-, negative control; lane 1, 4, 5, 7, 9, positive samples; lane 2, 3, 6, 8, negative samples.

General clinical symptoms in patients with atypical pneumonia

5. Discussion
Respiratory tract infections are major causes of global morbidity and mortality. Atypical pathogens cause about one-fifth of community-acquired pneumonia. Mycoplasma pneumoniae has a high prevalence worldwide. The role of M. pneumoniae in isolated human Mycoplasma is proven as a true pathogen in the respiratory tract (3, 24, 25). Atypical M. pneumoniae begins with nonspecific symptoms and is characterized by progression of symptoms of the upper respiratory tract to the lower one (3, 8). Rapid and accurate diagnosis is a factor that reduces mortality, morbidity, and costs of infectious diseases (26). In the present study, PCR was developed based on specific primers of M. pneumoniae and P1 gene. After collecting 102 clinical specimens from atypical pneumonia, the existence of P1 gene of M. pneumoniae was confirmed only in 14 specimens.
In a study by He et al. on 12025 children with respiratory infections, it was found that 20.23% of children had M. pneumoniae infection (2) that was more than the present study. In a study by Maheshwari et al. on 75 patients with a lower respiratory tract infection (30.7%) by PCR method, it was concluded that the incidence and prevalence of this disease were twice as much as the present research. In a study by Wu PS et al. in Taiwan in 2013 on 412 patients, the prevalence of M. pneumoniae was reported as 15% (27); and it was almost consistent with the present study. In a study by Reinton et al., the prevalence of atypical pneumonia (9.5%) was reported in 26039 Norwegian patients (28) and it was less frequent than the present study.
In the research by Medjo et al. who sought to determine the prevalence of M. pneumoniae infection in children with community-acquired pneumonia in 166 clinical samples, it was found that 14.5% of cases were afflicted with community-acquired pneumonia (29) that was almost consistent with the present study. According to a study in India, the prevalence of pneumonia by M. pneumoniae was reported as 7% (30) that was lower than the incidence of M. pneumoniae in the present study. Among approximately 500000 cases of community-acquired pneumonia, 20% were due to respiratory infections and should be hospitalized every year in the United States. Up to 35% of cases of outpatient pneumonia and up to 18% of cases of pneumonia requiring hospitalization were caused by M. pneumoniae (31). The incidence of atopic pneumonia caused by M. pneumoniae is different in various regions of Iran and low in most studies. In a study by Sharifi et al. with the aim to identify respiratory infections caused by M. pneumoniae in 200 cases in Tabriz, the prevalence of this bacterium was 6% (32), and it was consistent with the present study. In this study, the prevalence of M. pneumoniae infection was 13.13% that was lower than neighboring countries such as Iraq (19.4%) and Turkey (16.2%), and countries like Poland (52%), the United States (27% and 29.5%), Korea (40%) and Japan (24.2%). However, it was more prevalent than in some countries like India (7%) (30, 32).
5.1. Conclusions
Based on the research results, the prevalence of pneumonia caused by M. pneumoniae was 13.7% that was more than other regions of Iran such as Ahwaz, Rasht, and Tabriz. The research method provided precise results considering that all patients, who had positive M. pneumoniae species according to PCR molecular method, had clinical symptoms including the respiratory infection (such as weakness and lethargy, fatigue, persistent headache and dry cough, shortness of breath, diarrhea, sputum, and muscle ache). Based on results, the molecular PCR method if 16S rRNA gene relating to the Mycoplasma genus (32.4%) and P1 gene relating to M. pneumonia species (13.7%) were diagnosed rapidly and accurately in patients with respiratory infections; and these results indicated that in 18.7% of cases with 16S rRNA gene, the relevant species were unknown indicating that in addition to M. pneumoniae, other Mycoplasma species are present in the respiratory tract that should be diagnosed and taken into consideration. According to obtained results, the frequency of patients with respiratory infections due to M. pneumonia was 13.7% indicating that in addition to M. pneumonia, other bacterial agents and viral infections are involved in respiratory infections and should be diagnosed and taken into account. It was also found that the molecular PCR method was a quick and sensitive technique to diagnose M. pneumonia and had a higher sensitivity and specificity than other methods, especially the culture.
Acknowledgements
References
-
1.
Ke LQ, Wang FM, Li YJ, Luo YC. [Epidemiological characteristics of Mycoplasma pneumoniae pneumonia in children]. Zhongguo Dang Dai Er Ke Za Zhi. 2013;15(1):33-6. Chinese. [PubMed ID: 23336165].
-
2.
He XY, Wang XB, Zhang R, Yuan ZJ, Tan JJ, Peng B, et al. Investigation of Mycoplasma pneumoniae infection in pediatric population from 12,025 cases with respiratory infection. Diagn Microbiol Infect Dis. 2013;75(1):22-7. [PubMed ID: 23040512]. https://doi.org/10.1016/j.diagmicrobio.2012.08.027.
-
3.
Dash S, Chaudhry R, Dhawan B, Dey AB, Kabra SK, Das BK. Clinical spectrum and diagnostic yields of Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia. J Lab Physicians. 2018;10(1):44-9. [PubMed ID: 29403204]. [PubMed Central ID: PMC5784292]. https://doi.org/10.4103/JLP.JLP_62_17.
-
4.
Saraya T. Mycoplasma pneumoniae infection: Basics. J Gen Fam Med. 2017;18(3):118-25. [PubMed ID: 29264006]. [PubMed Central ID: PMC5689399]. https://doi.org/10.1002/jgf2.15.
-
5.
Meyer Sauteur PM, Unger WW, Nadal D, Berger C, Vink C, van Rossum AM. Infection with and Carriage of Mycoplasma pneumoniae in Children. Front Microbiol. 2016;7:329. [PubMed ID: 27047456]. [PubMed Central ID: PMC4803743]. https://doi.org/10.3389/fmicb.2016.00329.
-
6.
Page CA, Krause DC. Protein kinase/phosphatase function correlates with gliding motility in Mycoplasma pneumoniae. J Bacteriol. 2013;195(8):1750-7. [PubMed ID: 23396910]. [PubMed Central ID: PMC3624554]. https://doi.org/10.1128/JB.02277-12.
-
7.
Schmidl SR, Otto A, Lluch-Senar M, Pinol J, Busse J, Becher D, et al. A trigger enzyme in Mycoplasma pneumoniae: Impact of the glycerophosphodiesterase GlpQ on virulence and gene expression. PLoS Pathog. 2011;7(9). e1002263. [PubMed ID: 21966272]. [PubMed Central ID: PMC3178575]. https://doi.org/10.1371/journal.ppat.1002263.
-
8.
Xiao L, Ptacek T, Osborne JD, Crabb DM, Simmons WL, Lefkowitz EJ, et al. Comparative genome analysis of Mycoplasma pneumoniae. BMC Genomics. 2015;16:610. [PubMed ID: 26275904]. [PubMed Central ID: PMC4537597]. https://doi.org/10.1186/s12864-015-1801-0.
-
9.
Duffy MF, Whithear KG, Noormohammadi AH, Markham PF, Catton M, Leydon J, et al. Indirect enzyme-linked immunosorbent assay for detection of immunoglobulin G reactive with a recombinant protein expressed from the gene encoding the 116-kilodalton protein of Mycoplasma pneumoniae. J Clin Microbiol. 1999;37(4):1024-9. [PubMed ID: 10074521]. [PubMed Central ID: PMC88644].
-
10.
Shimizu T. Inflammation-inducing factors of Mycoplasma pneumoniae. Front Microbiol. 2016;7:414. [PubMed ID: 27065977]. [PubMed Central ID: PMC4814563]. https://doi.org/10.3389/fmicb.2016.00414.
-
11.
Kazama I, Nakajima T. Dual infection of Mycoplasma pneumoniae and Chlamydophila pneumoniae in patients with atopic predispositions successfully treated by moxifloxacin. Infez Med. 2014;22(1):41-7. [PubMed ID: 24651090].
-
12.
Wang ZH, Li XM, Wang YS, Guo ZY. Changes in the levels of interleukin-17 between atopic and non-atopic children with Mycoplasma pneumoniae pneumonia. Inflammation. 2016;39(6):1871-5. [PubMed ID: 27531365]. https://doi.org/10.1007/s10753-016-0422-3.
-
13.
Wang L, Feng Z, Zhao M, Yang S, Yan X, Guo W, et al. A comparison study between GeXP-based multiplex-PCR and serology assay for Mycoplasma pneumoniae detection in children with community acquired pneumonia. BMC Infect Dis. 2017;17(1):518. [PubMed ID: 28743259]. [PubMed Central ID: PMC5527399]. https://doi.org/10.1186/s12879-017-2614-3.
-
14.
Xu W, Guo L, Dong X, Li X, Zhou P, Ni Q, et al. Detection of viruses and Mycoplasma pneumoniae in hospitalized patients with severe acute respiratory infection in northern China, 2015-2016. Jpn J Infect Dis. 2018;71(2):134-9. [PubMed ID: 29491245]. https://doi.org/10.7883/yoken.JJID.2017.412.
-
15.
Kim JH, Cho TS, Moon JH, Kim CR, Oh JW. Serial changes in serum eosinophil-associated mediators between atopic and non-atopic children after Mycoplasma pneumoniae pneumonia. Allergy Asthma Immunol Res. 2014;6(5):428-33. [PubMed ID: 25229000]. [PubMed Central ID: PMC4161684]. https://doi.org/10.4168/aair.2014.6.5.428.
-
16.
Yu Y, Fei A. Atypical pathogen infection in community-acquired pneumonia. Biosci Trends. 2016;10(1):7-13. [PubMed ID: 26961211]. https://doi.org/10.5582/bst.2016.01021.
-
17.
Youn YS, Lee SC, Rhim JW, Shin MS, Kang JH, Lee KY. Early additional immune-modulators for Mycoplasma pneumoniae pneumonia in children: An observation study. Infect Chemother. 2014;46(4):239-47. [PubMed ID: 25566403]. [PubMed Central ID: PMC4285006]. https://doi.org/10.3947/ic.2014.46.4.239.
-
18.
Cilloniz C, Torres A, Niederman M, van der Eerden M, Chalmers J, Welte T, et al. Community-acquired pneumonia related to intracellular pathogens. Intensive Care Med. 2016;42(9):1374-86. [PubMed ID: 27276986]. https://doi.org/10.1007/s00134-016-4394-4.
-
19.
Sharma L, Losier A, Tolbert T, Dela Cruz CS, Marion CR. Atypical pneumonia: Updates on legionella, chlamydophila, and Mycoplasma pneumonia. Clin Chest Med. 2017;38(1):45-58. [PubMed ID: 28159161]. [PubMed Central ID: PMC5679202]. https://doi.org/10.1016/j.ccm.2016.11.011.
-
20.
Carrim M, Wolter N, Benitez AJ, Tempia S, du Plessis M, Walaza S, et al. Epidemiology and molecular identification and characterization of Mycoplasma pneumoniae, South Africa, 2012-2015. Emerg Infect Dis. 2018;24(3):506-13. [PubMed ID: 29460736]. [PubMed Central ID: PMC5823326]. https://doi.org/10.3201/eid2403.162052.
-
21.
Maheshwari M, Kumar S, Sethi GR, Bhalla P. Detection of Mycoplasma pneumoniae in children with lower respiratory tract infections. Trop Doct. 2011;41(1):40-2. [PubMed ID: 21123487]. https://doi.org/10.1258/td.2010.100149.
-
22.
Al-Ghizawi GJ, Al-Sulami AA, Al-Taher SS. Profile of community- and hospital-acquired pneumonia cases admitted to Basra General Hospital, Iraq. East Mediterr Health J. 2007;13(2):230-42. [PubMed ID: 17684843].
-
23.
Golmohammadi R, Ataee RA, Alishiri GH, Mirnejad R, Najafi A, Tate M, et al. [Molecular diagnosis of Mycoplasma pneumoniae in synovial fluid of rheumatoid arthritis patients]. Iran J Med Microbiol. 2014;8(1):1-8. Persian.
-
24.
Wijesooriya LI, Kok T, Perera J, Tilakarathne Y, Sunil-Chandra NP. Mycoplasma pneumoniae DNA detection and specific antibody class response in patients from two tertiary care hospitals in tropical Sri Lanka. J Med Microbiol. 2018;67(9):1232-42. [PubMed ID: 30074476]. https://doi.org/10.1099/jmm.0.000813.
-
25.
Schelonka RL, Waites KB. Ureaplasma infection and neonatal lung disease. Semin Perinatol. 2007;31(1):2-9. [PubMed ID: 17317421]. https://doi.org/10.1053/j.semperi.2007.01.001.
-
26.
Shahhosseiny M, Mardani M, Hosseini Z, Khorm Khorshid HR, Rahimi AA, Wand Yousefi J. [Comparison of culture and CPC tests for diagnose respiratory infections of Mycoplasma pneumoniae]. J Zanjan Univ. 2006;14(56):12-23. Persian.
-
27.
Wu PS, Chang LY, Lin HC, Chi H, Hsieh YC, Huang YC, et al. Epidemiology and clinical manifestations of children with macrolide-resistant Mycoplasma pneumoniae pneumonia in Taiwan. Pediatr Pulmonol. 2013;48(9):904-11. [PubMed ID: 23169584]. https://doi.org/10.1002/ppul.22706.
-
28.
Reinton N, Manley L, Tjade T, Moghaddam A. Respiratory tract infections during the 2011 Mycoplasma pneumoniae epidemic. Eur J Clin Microbiol Infect Dis. 2013;32(6):835-40. [PubMed ID: 23354674]. https://doi.org/10.1007/s10096-013-1818-8.
-
29.
Medjo B, Atanaskovic-Markovic M, Radic S, Nikolic D, Lukac M, Djukic S. Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia in children: clinical features and laboratory diagnosis. Ital J Pediatr. 2014;40:104. [PubMed ID: 25518734]. [PubMed Central ID: PMC4279889]. https://doi.org/10.1186/s13052-014-0104-4.
-
30.
Chaudhry R, Valavane A, Sreenath K, Choudhary M, Sagar T, Shende T, et al. Detection of Mycoplasma pneumoniae and Legionella pneumophila in patients having community-acquired pneumonia: A multicentric study from New Delhi, India. Am J Trop Med Hyg. 2017;97(6):1710-6. [PubMed ID: 29016299]. [PubMed Central ID: PMC5805046]. https://doi.org/10.4269/ajtmh.17-0249.
-
31.
Talkington DF, Thacker WL, Keller DW, Jensen JS. Diagnosis of Mycoplasma pneumoniae infection in autopsy and open-lung biopsy tissues by nested PCR. J Clin Microbiol. 1998;36(4):1151-3. [PubMed ID: 9542959]. [PubMed Central ID: PMC104711].
-
32.
Sharifi S, Ghottaslo R, Akhi MT, Soroush MH, Ansarin K, Shabanpour J. [Identification of respiratory infections caused by Mycoplasma pneumoniae With three methods of cultivation, ELISA and PCR]. Med J Tabriz Univ. 2011;23(3):36-41. Persian.