Abstract
Background:
The human cytomegalovirus (HCMV) is a common pathogen which usually remains asymptomatic in the healthy adults; however, it can cause a symptomatic disease in the immunocompromised patients. The risk of infection with HCMV increases in ulcerative colitis (UC) patients as a result of receiving immunosuppressive agents.Objectives:
This study aimed to determine the prevalence and the glycoprotein B genotypes of HCMV among the patients with HCMV disease superimposed on an UC flare that required hospitalization in Imam Khomeini Hospital in Ahvaz, Iran, during 2010- 2012.Patients and Methods:
In this case-control study, formalin-fixed paraffin-embedded intestinal tissue samples were taken from 98 patients with UC disease including 53 males and 45 females (mean age ± standard deviation, 38.95 ± 17.93) and 67 control patients with noninflammatory disease who were referred to Imam Khomeini Hospital during 2010-2012. Detection of HCMV genome in intestinal samples was carried out by seminested polymerase chain reaction. Glycoprotein B genotypes were determined by sequencing.Results:
Among 98 patients with UC, only 12 (12.2%) patients were positive for HCMV genome, while the HCMV genome was not detected in any of the controls. (P = 0.002). The distribution of HCMV gB genotypes in 12 CMV-positive UC patients was as follow: gB1, 11 (91.7%) and gB3, 1 (8.3%). The most prevalent genotype in CMV-positive UC patients was gB1.Conclusions:
In this study, high prevalence of 91.7% HCMV gB1 genotype was predominant among HCMV-positive UC patients, which suggests that there might be an association between HCMV gB genotype 1 and UC disease.Keywords
Human Cytomegalovirus Ulcerative Colitis gB Genotypes Nested PCR Intestinal Tissue
1. Background
Inflammatory bowel disease (IBD) is defined as chronic inflammatory disease of the gastrointestinal tract with several periods of clinical relapse and remission. There are two forms of IBD including Crohn’s disease (CD) and ulcerative colitis (UC) (1). Ulcerative colitis is usually more frequent than CD (2). It affects the colon and its symptoms are bloody diarrhea and rectal bleeding (3, 4). Crohn’s disease often affects the terminal ileum and colon with symptoms, such as abdominal pain, diarrhea and weight loss (1). Gastrointestinal cancer may be seen in both forms of IBD (1). Although it is said that several factors, including genetic and environmental, bacterial and viral, nutritional, psychological and immunological factors seem to be involved in causing IBD, the exact etiology of this disease has not been known yet (5). In this study, we focused on the viral factors. As we know, IBD has several periods of clinical relapse and remission and this feature is similar to infections caused by viruses of Herpesviridae family (1). Human cytomegalovirus (HCMV) is a double stranded DNA virus which belongs to the Herpesviridae family and is a beta human herpesvirus type 5 (6, 7). Human cytomegalovirus affects between 40% and 100% of the general population (6, 8); it usually remains asymptomatic in the healthy adults; however, it can cause symptomatic disease in the immunocompromised patients (7). Human cytomegalovirus disease may occur in various organs. HCMV infections involve any part of the gastrointestinal tract, and induce ulcers with hemorrhage (4). Recently, an association between IBD and the presence of HCMV has been reported (4, 9). The risk of infection with HCMV increases in IBD patients as a result of receiving immunosuppressive agents (10, 11), and this viral infection evoke the severity of inflammatory diseases (12, 13); however, the role of antiviral treatment is also unclear. To date, there haven’t been any controlled trials of antiviral therapy to clarify this issue (8); therefore, we cannot decide about the use of antiviral therapy in these patients (6). More studies are needed to confirm this.
Human cytomegalovirus shows genetic variations in virulent genes, such as UL55 (14, 15). The UL55 gene encodes glycoprotein B (gB), which is the major envelope glycoprotein of CMV (15, 16). On the basis of genetic variation in the UL55 gene, HCMV can be classified into 4 glycoprotein B (gB) genotypes (17). Also, the HCMV gB has an important role in tissue tropism, virus penetration into the host cell and fusion of infected cells. Therefore, it is an important target for humoral and cellular immune responses (15). Since the UC is recurrent and is one of the important causes for referring to the specialist, hospitalization of the patients accompany with work-loss and high treatment costs (3), an evaluation of rapid diagnosis of HCMV infection is important for an appropriate antiviral treatment and to prevent the serious complications of the disease. This study was conducted to detect HCMV DNA in intestinal tissue specimens of patients with flare UC by the nested polymerase chain reaction (PCR) in Ahvaz city, Iran. Ahvaz city is the center of Khuzestan province located in the south-west of Iran with 1.5 million populations.
2. Objectives
This study aimed to determine the prevalence and the glycoprotein B genotypes of HCMV among the patients with HCMV disease superimposed on an UC flare that required hospitalization in Imam Khomeini Hospital in Ahvaz, Iran, during 2010- 2012.
3. Patients and Methods
3.1. Patients and Samples
Intestinal samples were taken from 98 patients with UC disease including 53 males and 45 females (mean age ± SD, 38.95 ± 17.93) referred to Imam Khomeini Hospital in Ahvaz city, Iran, during 2010 - 2012. All patients were diagnosed with UC by clinical, laboratory, endoscopic, and histological findings in accordance with Leonard-Jones criteria (18). The control group consisted of 67 individuals with noninflammatory disease, who were evaluated by colonoscopy for anemia, change bowel habit or rectal bleeding, granolomatosis or colonal papilomatosis diseases, and were matched in age (± 5 years), sex, and date of hospital admission with the UC patients. A written informed consent was obtained from all patients as well as control subjects.
3.2. Molecular Study
3.2.1. Preparation of Formalin-Fixed, Paraffin-Embedded Colon Tissues for DNA Extraction
Tissue sections (20 μm thick; 8 or 10 sections from each block) were deparaffinized with 1 mL xylene in 1.5 mL DNase-free micro tubes at 45˚C for 15 minutes. After 15 minutes, xylene was removed and this step was repeated twice. The tissues were washed with ethanol 100%, 80%, 60% and then ethanol 40%. The tubes were kept at room temperature with an opened cap to completely evaporate ethanol.
3.2.2. DNA Extraction
The DNA extraction from deparaffinized colon specimens was performed by the high- pure PCR template preparation kit (Roche, Germany) according to the manufacturer’s instructions. The extracted DNA was eluted in 100 μL DNase-free Mili-Q water and stored at -20˚C before being used as a template to detect CMV DNA.
3.2.3. Seminested Polymerase Chain Reaction Assay
Human cytomegalovirus DNA was detected using the seminested PCR with the paired primers conserved for the gB gene (UL55 region) including, gB1043/gB 1724 and gB1043/gB1604 (15, 19). The sequences of PCR primers used in this study are listed in Table 1.
Nested PCR Primer Sequences
| Primer | Sequence | Primer Pair | Product Size |
|---|---|---|---|
| gB1043 | (nt 1043-1061): 5-TCTGGGAAGCCTCGGAACG-3 | gB1043/gB 1724 | 680 - 685 bp |
| gB1724 | (nt 1724-1704): 5-GAGTAGCAGCGTCCTGGCGA-3 | ||
| gB1604 | (nt 1621-1604): 5-GAAACGCGCGGCAATCGG-3 | gB1043/gB1604 | 580 - 583 bp |
Human beta-globin gene was used as internal control to check the absence of PCR inhibitors and DNA quality (20). DNA quality was evaluated by PCR using forward primer PCO3: 5’-ACACAACTGTGTTCACTAGC-3’ and reverse primer PCO4: 5’-CAACTTCATCCACGTTCACC-3’ that amplify a 110 bp product from the human β-globin gene (21). The first PCR was conducted in a total volume of 25 μL of a reaction buffer that included the following components: 0.4 μM of each gB1043 and gB1724 outer primer (10 pmol/reaction), 200 μM of each of the 4 deoxynucleotides, 100 ng DNA template, 1.25 U of Taq DNA polymerase, 1 × PCR buffer containing 1.5 mmol/L MgCl2 and DNase-free Mili-Q water. PCR components were purchased from Roche Company (Germany).
Themocycler (TC-512, Techne, UK) was programmed with an initial denaturation for 5 minutes at 95˚C, and then followed by 35 cycles consisting of a denaturation step, 94˚C for 40 seconds, an annealing step, 57˚C for 50 seconds, and an extension step was 72˚C for 60 seconds. The final extension cycle was 72˚C for 5 minutes. For the second reaction, 2 μL of the first PCR product was added to a tube containing the 0.4 µM of each gB1043 and gB1604 inner primers (10 pmol/reaction), with the same PCR reagents as in the first reaction. Second thermocycling conditions were as follows: Initial denaturation at 95˚C for 5 minutes, 35 cycles consisting of 94˚C for 40 seconds, 58˚C for 50 seconds, 72˚C for 60 seconds, and 72˚C for 5 minutes. PCR products were analyzed by electrophoresis on 2% agarose gel (Cinagene, Iran) containing DNA Safe Stain and electrophoresed for about 45 minutes at 100 volts in 0.5 × TBE buffer. The gel was visualized on UV Transilluminater (Vilber Lourmat, France). A DNA band with ≈ 580 bp was considered as a positive sample and subjected to genotyping. The sequence of both strands was determined.
3.2.4. DNA Sequencing Assay
The CMV-positive PCR products were purified with a QIAQuick PCR purification kit (Qiagen, United Kingdom) and were subjected for automated sequencing (Takapozist Co., Tehran, Iran). Nucleotide sequences of CMV strains were aligned using ClustalW (22).
3.3. Nucleotide Sequence Accession Numbers and Phylogenetic Analysis
Genotyping of HCMV glycoprotein B (gB) was carried out by sequencing for 12 CMV-positive samples. The Phylogenetic tree analysis was used to compare the genetic distances for all 12 isolates. The phylogenetic tree was constructed by the neighbor-joining method with the Bootstrap test of phylogeny in Molecular Evolutionary Genetics Analysis (MEGA) program, version 4 (23, 24). Bootstrap resampling strategy and reconstruction were carried out 1000 times to confirm the reliability of the phylogenetic tree (25).
3.4. Statistical Analysis
All data were presented as mean ± SD or frequencies. Clinical characteristics were compared using the chi-square test or Fisher’s exact test and Student’s t-test. Data were processed by SPSS for statistical analysis (SPSS Inc., Chicago, IL) and P-values of 0.05 or less reported as statistically significance.
4. Results
A total of 98 cases with flare UC, including 53 males and 45 females (mean age ± SD, 38.95 ± 17.93), referred to Imam Khomeini Hospital in Ahvaz city during 2010-2012, and 67 controls with noninflammatory disease, who were matched in age (± 5 years), sex, and date of hospital admission with UC patients, were participated in this study. The common intestinal locations for UC were proctitis (13.3%), pancolitis (19.4%), rectosigmoid (21.4%), left-side colitis (11.2%), rectosigmoid-decending colon (18.4%) and extend colitis (16.3%). The highest rate of UC was observed in rectosigmoid and pancolitis, whereas the lowest rate was in left colitis. Sixty-one (62%) flare UC patients were treated with immunosuppresive drugs during sampling. The duration of flare UC disease was 4.53 ± 3.54 years. The demographic and clinical profiles of the 98 flare UC patients are summarized in Table 2. The histopathologic examination confirmed diagnosis of UC in all patients of the case group; whereas the absence of any inflammatory disease was confirmed in the control group. The β-globin DNA was detectable in all formalin-fixed paraffin-embedded specimens.
Demographic and Clinical Parameters of Patients Included in the Study
| Characteristics and Parameters | Values |
|---|---|
| Age, y | 38.95 ± 17.93 a |
| Duration of Disease, y | 4.53 ± 3.54 a |
| Gender b | |
| Male | 53 (54.1) |
| Female | 45 (45.9) |
| Intestinal Location b | |
| Proctitis | 13 (13.3) |
| Pancolitis | 19 (19.4) |
| Rectosigmoid | 21 (21.4) |
| Left-sided colitis | 11 (11.2) |
| Rectosigmoid-decending colon | 18 (18.4) |
| Up to transverse colon | 16 (16.3) |
| Immunosuppressive Drug User b | |
| No | 37 (38) |
| Yes | 61 (62) |
| Race b | |
| Persian | 69 (70.4) |
| Arabic | 29 (29.6) |
| Place b | |
| Urban | 89 (90.8) |
| Rural | 9 (9.2) |
| Education | |
| Diploma and post diploma | 62 (63.3) |
| Under diploma | 36 (36.7) |
| Other Sick | |
| Anemia | 16 (16.3) |
| Hypertension | 5 (5.1) |
| PSC | 8 (8.2) |
| Thyroid disorder | 9 (9.2) |
| Diabetes | 6 (6.1) |
| Hepatitis | 3 (3.1) |
| None | 51 (52) |
The HCMV genome in the intestinal tissue was found in 12 (12.2%) UC patients. Figure 1 shows the ≈ 580bp PCR product. All of the biopsy samples of the control group were negative for HCMV. These 12 CMV-positive UC patients were 8 males (66.7%) and 4 females (33.3%) with mean age ± SD of 35.17 ± 9.85. The highest rate of the intestinal location of UC was in pancolitis (41.7%). Table 3 summarizes the association of various demographic and clinical variables associated with CMV-infection. In this study, detection of CMV genome in the intestine was commonly associated with UC (P = 0.002). However, detection of CMV genome was not statistically associated with age and sex, duration of disease, race, place, education, location of disease, and use of immunosuppressive drugs.
The Nested Polymerase Chain Reaction Amplification of Human Cytomegalovirus DNA From Biopsy Sample
Demographic and Clinical Data for Cytomegalovirus -Positive and Cytomegalovirus -Negative UC Patients
| Characteristics and parameters | CMV-positive (n = 12) | CMV-negative (n = 86 ) | P value |
|---|---|---|---|
| Age, y a | 35.17 ± 9.85 | 39.48 ± 18.76 | 0.229 |
| Duration of disease, y a | 3.27 ± 2.74 | 4.70 ± 3.61 | 0.192 |
| Gender b | |||
| Male | 8 (66.7) | 45 (52.3) | 0.35 |
| Female | 4 (33.3) | 41 (47.2) | |
| Intestinal locationb | |||
| Proctitis | 2 (16.7) | 11 (12.8) | 0.351 |
| Pancolitis | 5 (41.7) | 14 (16.3) | |
| Rectosigmoid | 1 (8.3) | 20 (23.3) | |
| Left-sided colitis | 1 (8.3) | 10 (11.6) | |
| Rectosigmoid-decending colon | 1 (8.3) | 17 (19.8) | |
| Up to transverse colon | 2 (16.7) | 14 (16.3) | |
| Immunosuppressive Drug Userb | |||
| No | 5 (41.7) | 32 (37.2) | 0.765 |
| Yes | 7 (58.3) | 54 (62.8) | |
| Race b | |||
| Persian | 6 (50) | 63 (73.3) | 0.098 |
| Arabic | 6 (50) | 23 (26.7) | |
| Placeb | |||
| Urban | 10 (83.3) | 79 (91.9) | 0.338 |
| Rural | 2 (16.7) | 7 (8.1) | |
| Education | |||
| Diploma and post diploma | 8 (66.7) | 54 (62.8) | 0.798 |
| Under diploma | 4 (33.3) | 32 (37.2) | |
| Other Sick | |||
| Anemia | 2 (16.7) | 14 (16.3) | 0.273 |
| Hypertension | 1 (8.3) | 4 (4.7) | |
| PSC | 3 (25) | 5 (5.8) | |
| Thyroid disorder | 0 (0.0) | 9 (10.5) | |
| Diabetes | 0 (0.0) | 6 (6.9) | |
| Hepatitis | 0 (0.0) | 3 (3.5) | |
| None | 6 (50) | 45 (52.3) |
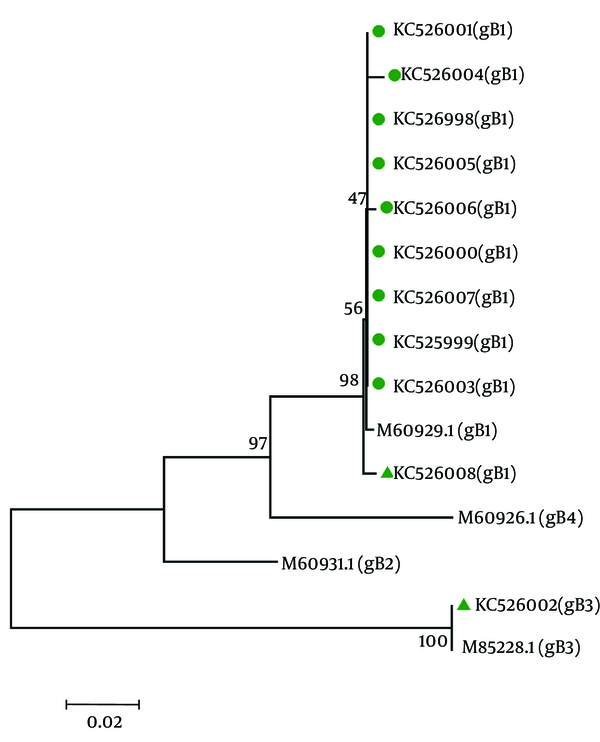
The GenBank accession numbers for the HCMV sequences isolated from CMV-positive UC patients in this article are KC525998, KC525999, KC526000, KC526001, KC526002, KC526003, KC526004, KC526005, KC526006, KC526007, and KC526008. Figure 2 shows the trees generated by using sequences from gB gene region. The result of CMV-genotyping by sequencing based on MEGA software revealed 11 cases (91.7%) for gB1, and 1 case (8.3%) for gB3 genotypes, respectively among the UC patients. The most prevalent genotype in HCMV-positive flare UC patients was gB1.
Phylogenetic Tree Based on gb Gene Sequences of Cytomegalovirus-Isolates From Intestinal Biopsy of 12 Cytomegalovirus -Positive Ulcerative Colitis Patients During 2010 and 2012
5. Discussion
The human cytomegalovirus is a common pathogen, which occurs early in life. It usually causes asymptomatic infections and remains persistent in healthy individuals (9), but it causes symptomatic diseases in immunocompromised patients. In these patients, the HCMV disease may occur in various organs (7). In recent years, the role of HCMV disease in IBD has been described (26, 27) and HCMV tends to invade and stay in inflammatory areas (28). Orvar et al. proposed that the cross-reactive immune responses between human and viral proteins might cause an autoimmune response, which lead to IBD (29). On the other hand, IBD patients are in immunodeficiency state as a result of treating with immunosuppressive agents; therefore, the risk of HCMV infection increases in these patients and this virus can worsen the condition of IBD (10, 11). Thus, HCMV infection might cause UC or occur as a result of UC injuries in the intestinal mucosa. The exact role of HCMV in UC disease is still unclear (28).
In recent years, some studies have reported the high prevalence of HCMV in intestinal tissue biopsies among IBD patients in different regions of the world (9, 28), and even CMV cytotoxic auto antibodies have been detected in more than 50% of patients with either UC or CD (30, 31). Criscuoli et al. discovered CMV infection prevalence of 21.4% among IBD patients in Italy (32). In another survey, Kandiel et al. studied the prevalence of CMV infection in patients with IBD. In their study, CMV infection prevalence was 21%- 34% among American patients (12). In the present study, the HCMV prevalence was evaluated in flare UC patients in comparison with individuals with noninflammatory disease as control group. In the intestinal biopsy, CMV genome was detected in 12.2% of UC patients but not detected in the control group. Recently, some studies have reported that there is an association between CMV infection and IBD (10, 28, 32).
In our study, an association was observed between the presence of CMV in the intestinal biopsy and UC disease. However, no significant difference was found between CMV-positive UC patients and CMV-negative UC patients regarding several demographic and clinical parameters. Cottone et al. studied the prevalence of CMV infection among IBD patients in Italy. In their study, CMV was diagnosed in the rectal specimens of seven (five with UC and two with CD) out of 19 (36%) patients with refractory disease. They came to this conclusion that the CMV infection is a frequent cause of IBD (26). In another survey, Wada et al. detected CMV infection in 16 of 47 patients (34%) with UC in Japan. Proportion of female patients, age at the time of determination was significantly higher in the CMV-positive group (59.1%) than in the CMV-negative group (13.6%). The prevalence of endoscopically severe UC was higher in patients with CMV antigenemia than in those without CMV antigenemia (P = 0.016) (27).
Dimitroulia et al. studied frequent detection of CMV in the intestine of patients with IBD in Greece. In their study, CMV genome was found in 27.1% of the IBD patients, while in the control group was 2.2%. Since the CMV infection was frequently found in patients with IBD as compared with the control group, they came to this conclusion that detection of CMV genome in the intestine was associated with IBD (9). Although there was an association between HCMV and UC disease, the exact role of CMV in IBD patients is still unclear; some studies suggest that CMV has a nonpathogenic role in the intestine of IBD patients, while others report the adverse effects of CMV on clinical results of IBD patients (13, 32, 33). This diversity of clinical outcome of CMV might be explained by studies on the genotyping of the virus.
In previous studies, the gB genotype 1 and gB genotype 2 were found more frequently in different groups such as bone marrow, stem cell and renal transplant recipients, immunocompromised and HIV infected patients (16, 34, 35). However, among gB type 1 and type 2, gB type 1 was the most frequent (16), while type 2 was more frequent in HIV-infected patients (35, 36). Some studies revealed that mixed gB genotype infections are associated with higher viral loads, delayed viral clearance, virulence patterns, virologic outcomes, progression to CMV disease, increased rate of graft rejection, more often coinfections with other herpesviruses and sever clinical manifestation (14, 16, 37). In our study, the result of CMV genotyping revealed 11 (91.7%) for gB1, and 1 (8.3%) for gB3 genotypes, respectively among the UC patients. In present study, HCMV gB1 genotype was predominant, which suggests that there might be an association between gB genotype 1 and UC disease. More investigation is needed to confirm this.
Regarding HCMV genotypes, Coaquette et al. found gB1 (28.9%) , gB2 (19.6%), gB3 (23.7%); gB4 (2.0%) and mixed genotypes in 25.8% among immunocompromised patients, and revealed that the presence of multiple gB genotypes in infected patients associated with severe clinical outcomes in contrast to patients infected with a single gB genotype (16). Gonzalez-Ramirez et al. studied the frequency distribution of CMV genotypes in Mexican children with allogeneic bone-marrow transplantation. They found gB1 (30%), gB2 (27%), gB3 (13%), gB4 (3%) and mixed genotypes in 8/30 patients (27%). In their study, genotypes gB2 and gB1 had the highest frequency (34). In another study, Dieamant et al. determined the distribution of gB genotypes in allogeneic hematopoietic stem-cell transplant (HSCT) recipients with CMV infection in Brazil. They found gB1 (46.6%), gB2 (33.3%), gB3 (6.7%); gB4 (6.7%) and mixed infection (6.7%) genotypes in this population. In their study, genotype gB type 1 was the most prevalent genotype, and they revealed that the mixture of HCMV gB genotypes was associate with gastrointestinal disease (17).
Genotype information is important for predicting tropism and virulence of HCMV, treatment and antiviral resistance, progression to HCMV disease, graft rejection, viral loads and recurrent infection (14, 38). Since glycoprotein B (gB) is involved in virulence, virulence differs among different CMV genotypes (15, 38). Moreover, genotype determines the length of therapy e.g. mixed gB genotype infections are associated with higher viral loads and delayed viral clearance (14, 37). In addition, for treatment purposes, detection of HCMV genotypes in different regions and different groups of patients can be used for the purpose of molecular epidemiology (14). Furthermore, these findings can facilitate the choice of recombinant HCMV glycoprotein vaccine required to immunize high-risk group (39).
In conclusion, in this case-control study, overall CMV prevalence was 12.2% among the UC patients (P = 0.002). In present study, high prevalence of 91.7% CMV gB1 genotype was observed among the flare UC patients, which suggests that there might be an association between the gB genotype 1 and UC disease. However, further studies are needed to confirm this.
Acknowledgements
References
-
1.
Blumberg RS, Strober W. Prospects for research in inflammatory bowel disease. JAMA. 2001;285(5):643-7. [PubMed ID: 11176874].
-
2.
Sood A, Midha V, Sood N, Bhatia AS, Avasthi G. Incidence and prevalence of ulcerative colitis in Punjab, North India. Gut. 2003;52(11):1587-90. [PubMed ID: 14570727].
-
3.
Kornbluth A, Sachar DB, Practice Parameters Committee of the American College of G. Ulcerative colitis practice guidelines in adults (update): American College of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol. 2004;99(7):1371-85. [PubMed ID: 15233681]. https://doi.org/10.1111/j.1572-0241.2004.40036.x.
-
4.
Suzuki H, Kato J, Kuriyama M, Hiraoka S, Kuwaki K, Yamamoto K. Specific endoscopic features of ulcerative colitis complicated by cytomegalovirus infection. World J Gastroenterol. 2010;16(10):1245-51. [PubMed ID: 20222169].
-
5.
Sands BE, Grabert S. Epidemiology of inflammatory bowel disease and overview of pathogenesis. Med Health R I. 2009;92(3):73-7. [PubMed ID: 19385381].
-
6.
Al-Zafiri, R, Gologan, A.Z, Galiatsatos, P, Szilagyi, A. Cytomegalovirus complicating inflammatory bowel disease: a 10-year experience in a community-based, university-affiliated hospital. Gastroenterol Hepatol. 2012;8(4):230-9.
-
7.
Crough T, Khanna R. Immunobiology of human cytomegalovirus: from bench to bedside. Clin Microbiol Rev. 2009;22(1):76-98. Table of Contents. [PubMed ID: 19136435]. https://doi.org/10.1128/CMR.00034-08.
-
8.
Lawlor G, Moss AC. Cytomegalovirus in inflammatory bowel disease: pathogen or innocent bystander? Inflamm Bowel Dis. 2010;16(9):1620-7. [PubMed ID: 20232408]. https://doi.org/10.1002/ibd.21275.
-
9.
Dimitroulia E, Spanakis N, Konstantinidou AE, Legakis NJ, Tsakris A. Frequent detection of cytomegalovirus in the intestine of patients with inflammatory bowel disease. Inflamm Bowel Dis. 2006;12(9):879-84. [PubMed ID: 16954807]. https://doi.org/10.1097/01.mib.0000231576.11678.57.
-
10.
Kishore J, Ghoshal U, Ghoshal UC, Krishnani N, Kumar S, Singh M, et al. Infection with cytomegalovirus in patients with inflammatory bowel disease: prevalence, clinical significance and outcome. J Med Microbiol. 2004;53(Pt 11):1155-60. [PubMed ID: 15496396].
-
11.
Kim YS, Kim YH, Kim JS, Cheon JH, Ye BD, Jung SA, et al. Cytomegalovirus infection in patients with new onset ulcerative colitis: a prospective study. Hepatogastroenterology. 2012;59(116):1098-101. [PubMed ID: 22580660]. https://doi.org/10.5754/hge10217.
-
12.
Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol. 2006;101(12):2857-65. [PubMed ID: 17026558]. https://doi.org/10.1111/j.1572-0241.2006.00869.x.
-
13.
Hommes DW, Sterringa G, van Deventer SJ, Tytgat GN, Weel J. The pathogenicity of cytomegalovirus in inflammatory bowel disease: a systematic review and evidence-based recommendations for future research. Inflamm Bowel Dis. 2004;10(3):245-50. [PubMed ID: 15290919].
-
14.
Lisboa LF, Tong Y, Kumar D, Pang XL, Asberg A, Hartmann A, et al. Analysis and clinical correlation of genetic variation in cytomegalovirus. Transpl Infect Dis. 2012;14(2):132-40. [PubMed ID: 22093996]. https://doi.org/10.1111/j.1399-3062.2011.00685.x.
-
15.
Humar A, Kumar D, Gilbert C, Boivin G. Cytomegalovirus (CMV) glycoprotein B genotypes and response to antiviral therapy, in solid-organ-transplant recipients with CMV disease. J Infect Dis. 2003;188(4):581-4. [PubMed ID: 12898447]. https://doi.org/10.1086/377002.
-
16.
Coaquette A, Bourgeois A, Dirand C, Varin A, Chen W, Herbein G. Mixed cytomegalovirus glycoprotein B genotypes in immunocompromised patients. Clin Infect Dis. 2004;39(2):155-61. [PubMed ID: 15307021]. https://doi.org/10.1086/421496.
-
17.
Dieamant, DC, Bonon, SHA, Murakami, MM, Aranha, FJP, Duarte, GO, Fernandes, VCA. Genotyping of Human Cytomegalovirus (HCMV) Glycoprotein B (gB) in Hematopoietic Stem Cell Transplant Recipients with Active HCMV Infection: Impact of gB Genotypes on the Patient's Outcome. Biol. Blood. Marrow. Tr. 2011;17(2):315.
-
18.
Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2-6. discussion 16-9. [PubMed ID: 2617184].
-
19.
Gilbert C, Handfield J, Toma E, Lalonde R, Bergeron MG, Boivin G. Human cytomegalovirus glycoprotein B genotypes in blood of AIDS patients: lack of association with either the viral DNA load in leukocytes or presence of retinitis. J Med Virol. 1999;59(1):98-103. [PubMed ID: 10440815].
-
20.
Muciaccia B, Corallini S, Vicini E, Padula F, Gandini L, Liuzzi G, et al. HIV-1 viral DNA is present in ejaculated abnormal spermatozoa of seropositive subjects. Hum Reprod. 2007;22(11):2868-78. [PubMed ID: 17855413]. https://doi.org/10.1093/humrep/dem288.
-
21.
Sankaranarayanan R, Budukh AM, Rajkumar R. Effective screening programmes for cervical cancer in low- and middle-income developing countries. Bull World Health Organ. 2001;79(10):954-62. [PubMed ID: 11693978].
-
22.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-80. [PubMed ID: 7984417].
-
23.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406-25. [PubMed ID: 3447015].
-
24.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9. [PubMed ID: 17488738]. https://doi.org/10.1093/molbev/msm092.
-
25.
Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39(4):783. https://doi.org/10.2307/2408678.
-
26.
Cottone M, Pietrosi G, Martorana G, Casa A, Pecoraro G, Oliva L, et al. Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn's colitis. Am J Gastroenterol. 2001;96(3):773-5. [PubMed ID: 11280549]. https://doi.org/10.1111/j.1572-0241.2001.03620.x.
-
27.
Wada Y, Matsui T, Matake H, Sakurai T, Yamamoto J, Kikuchi Y, et al. Intractable ulcerative colitis caused by cytomegalovirus infection: a prospective study on prevalence, diagnosis, and treatment. Dis Colon Rectum. 2003;46(10 Suppl):S59-65. [PubMed ID: 14530660]. https://doi.org/10.1097/01.DCR.0000087486.21981.C6.
-
28.
Mariguela VC, Chacha SG, Cunha Ade A, Troncon LE, Zucoloto S, Figueiredo LT. Cytomegalovirus in colorectal cancer and idiopathic ulcerative colitis. Rev Inst Med Trop Sao Paulo. 2008;50(2):83-7. [PubMed ID: 18488086].
-
29.
Orvar K, Murray J, Carmen G, Conklin J. Cytomegalovirus infection associated with onset of inflammatory bowel disease. Dig Dis Sci. 1993;38(12):2307-10. [PubMed ID: 8261839].
-
30.
Rahbar A, Bostrom L, Lagerstedt U, Magnusson I, Soderberg-Naucler C, Sundqvist VA. Evidence of active cytomegalovirus infection and increased production of IL-6 in tissue specimens obtained from patients with inflammatory bowel diseases. Inflamm Bowel Dis. 2003;9(3):154-61. [PubMed ID: 12792220].
-
31.
Rahbar A, Bostrom L, Soderberg-Naucler C. Detection of cytotoxic CD13-specific autoantibodies in sera from patients with ulcerative colitis and Crohn's disease. J Autoimmun. 2006;26(3):155-64. [PubMed ID: 16584867]. https://doi.org/10.1016/j.jaut.2006.02.003.
-
32.
Criscuoli V, Casa A, Orlando A, Pecoraro G, Oliva L, Traina M, et al. Severe acute colitis associated with CMV: a prevalence study. Dig Liver Dis. 2004;36(12):818-20. [PubMed ID: 15646428]. https://doi.org/10.1016/j.dld.2004.05.013.
-
33.
Matsuoka K, Iwao Y, Mori T, Sakuraba A, Yajima T, Hisamatsu T, et al. Cytomegalovirus is frequently reactivated and disappears without antiviral agents in ulcerative colitis patients. Am J Gastroenterol. 2007;102(2):331-7. [PubMed ID: 17156136]. https://doi.org/10.1111/j.1572-0241.2006.00989.x.
-
34.
Gonzalez-Ramirez J, Uribe-Gutierrez G, Jimenez-Hernandez E, Velazquez-Guadarrama N, Bello-Gonzalez A, Vazquez-Meraz E, et al. Cytomegalovirus gB genotype distribution in Mexican children undergoing allogeneic bone marrow transplantation. Intervirology. 2012;55(4):318-20. [PubMed ID: 21734356]. https://doi.org/10.1159/000328665.
-
35.
de Campos Dieamant D, Bonon SH, Prates LC, Belangelo VM, Pontes ER, Costa SC. Active human cytomegalovirus infection and glycoprotein b genotypes in brazilian pediatric renal or hematopoietic stem cell transplantation patients. Braz J Microbiol. 2010;41(1):50-8. [PubMed ID: 24031463]. https://doi.org/10.1590/S1517-838220100001000010.
-
36.
Drew WL, Chou S, Miner RC, Mohr BA, Busch MP, van der Horst CM, et al. Cytomegalovirus glycoprotein B groups in human immunodeficiency virus-infected patients with incident retinitis. J Infect Dis. 2002;186(1):114-7. [PubMed ID: 12089671]. https://doi.org/10.1086/341071.
-
37.
Manuel O, Asberg A, Pang X, Rollag H, Emery VC, Preiksaitis JK, et al. Impact of genetic polymorphisms in cytomegalovirus glycoprotein B on outcomes in solid-organ transplant recipients with cytomegalovirus disease. Clin Infect Dis. 2009;49(8):1160-6. [PubMed ID: 19751151]. https://doi.org/10.1086/605633.
-
38.
Meyer-Konig U, Vogelberg C, Bongarts A, Kampa D, Delbruck R, Wolff-Vorbeck G, et al. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J Med Virol. 1998;55(1):75-81. [PubMed ID: 9580889].
-
39.
Woo PC, Lo CY, Lo SK, Siau H, Peiris JS, Wong SS, et al. Distinct genotypic distributions of cytomegalovirus (CMV) envelope glycoprotein in bone marrow and renal transplant recipients with CMV disease. Clin Diagn Lab Immunol. 1997;4(5):515-8. [PubMed ID: 9302197].