Abstract
Background:
Hepatitis C virus (HCV) causes one of the major chronic liver diseases (CLD). Hepatitis C virus- core encoding sequence possesses an overlapping open reading frame (ORF) that expresses a protein called F or core+1.Objectives:
The current study aimed at assessing the presence and titer of anti-core+1 antibody (Ab) in 70 Iranian patients infected with HCV-1a, responder and non-responder groups, under combination therapy with pegylated interferon-α (PegIFN-α) plus ribavirin (RBV) using an enzyme-linked immunosorbent assay (ELISA).Methods:
In the current cohort study, HCV-1a core+1 gene was amplified and cloned into vector followed by expressing in Escherichia coli and then, purified by ion exchange chromatography. The antibody titer of patients was evaluated before, during (12, 24, and 48 weeks), and 6 months after the end of therapy (ETR).Results:
The seroprevalence of anti-core+1 Ab was 75.7% in pretreatment sera. The combination therapy could induce a decline in the level of anti-core+1 Ab in both groups of responders and non-responders. These changes were significant only in the responders (P = 0.003). The seroprevalence of anti-core+1 Ab had no correlation with the outcome of treatment.Conclusions:
According to the current study results, HCV core+1 protein elicit a specific antibody response other than the anti-core protein antibodies. The current study data also suggested that the level of anti-core+1 antibody might be affected by the combination therapy and associated with sustained virological response (SVR). The data implied that the declining trend of anti-core+1 Abs during the treatment might be an alternative representation of the therapeutic response in Iranian population infected with HCV.Keywords
1. Background
Hepatitis C virus (HCV) is an enveloped RNA virus with a 9.6-kb genome classified in the genus Hepacivirus of the family Flaviviridae (1-3). Hepatitis C virus is one of the leading causes of persistent infection in humans that can predispose to liver fibrosis and cirrhosis. There are some reports indicating that chronic hepatitis C (CHC) infection is strongly associated with hepatocellular carcinoma (HCC) (3-9). There is neither effective vaccine for immunization, nor a broadly effective treatment for HCV infection (3).Among different factors associated with chronic HCV infection, alternate reading frame protein (ARFP) or core+1 protein is one the newest ones that can promote persistent infection (10-12). This protein is produced by a frame shift at codon 11 by a minority of ribosomes that initiate translation at the HCV AUG initiator codon of HCV genome (13, 14). Since this discovery, the specific functional and pathogenic roles of the core+1 protein still remain unclear (15).
Similar to some other HCV proteins, the core+1 protein may disturb signal pathway of the host cells leading to cellular abnormality and pathology (15). It is shown that core+1 protein interacts with p21, leading to inhibition of the cell cycle (16). Cohen et al., (5) suggested no significant correlation between the presence of anti-core+1 antibody (Ab) and HCV viremia or antiviral therapy outcome, while Gao et al., (17) found a correlation between anti-core+1 Ab and the outcome of treatment in patients infected with HCV. Many studies suggested the critical role of core+1 protein in humoral response, HCV infection status (12, 18) as well as outcomes (11, 19).
The detection of high levels of specific anti-core+1 antibodies in patients infected with HCV and HCC provides strong evidence that core+1 is produced during natural infection and may have a role in the progress to advanced stages of HCV liver disease (1, 12, 19, 20). In the study by Park et al. (21), it was shown that the expression of core+1 can lead to suppression of types I and III interferon (IFN) responses in hepatocytes, suggesting the important role of the core+1 in the modulation of host innate immunity by HCV.
2. Objectives
The current study aimed at characterizing the prevalence, titer dynamics of specific anti-core+1 Ab response, and its association with virological response in patients with CHC treated with combination therapy in both groups of responders and non-responders.
3. Methods
3.1. Ethics Statement
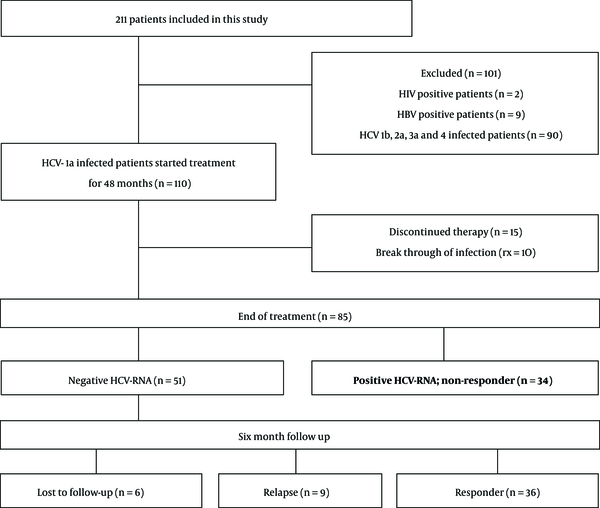
The study protocol was approved by the digestive disease research institute, institutional research board, and ethics committee (11 October, 2012; approval code: 416/501). The subjects provided informed consent for the use of their blood samples for research studies. The selection methodology is shown in Figure 1.
Flow Diagram of Included/Excluded Studies
3.2. Patients
In the current cohort study, 211 out-patients were visited in the clinic of a referral university hospital (Shariati Hospital, Tehran, Iran). The inclusion criteria were IFN-naïve patients with CHC and a negative antibody against HIV and HBV infections. None of the patients were at the cirrhosis stage, but histologically proven for chronic active hepatitis with positive results of polymerase chain reaction (PCR) for HCV RNA. Exclusion criteria were the liver disease of origin other than HCV infection and infection with another genotype of HCV (1b, 2a, 3a, 4). Finally, 70 adult patients with HCV genotype 1a infection were enrolled. The control group comprised of 30 adult Iranian healthy individuals (15 male, 15 female).
3.3. Recombinant core+1 Production, Purification, Western Blotting, and the Enzyme-Linked Immunosorbent Assay
The full-length of HCV-core 1a proteins was gifted by Dr. Jean-Pierre Lavergne from French national centre for scientific research, Paris, France (6). HCV core-1a cDNA sequences obtained from a plasmid (kindly provided by Dr. M Baril, departement of biochemistry, university of Montreal, Canada; based on the protocol of Baril et al. 2005) was used as the template for the amplification of core+1 gene as described elsewhere (7). Briefly, the cDNA fragment of the HCV core+1 protein coding sequence was achieved by +1/-1 frame-shifting artificially introduced at codon 31 of the core protein-coding sequence using the following design-specific primers:
Forward: 5-ATGAGCACGAATCCTAAACCTCAAAGAAAACC-3,
Reverse: 5-TAGTTCACGCCGTCTTCCAGAACC-3).
DNA fragments covering the complete core+1 sequence corresponding to amino acids 1 to 191 were amplified and cloned into the pET-28a (+) expression vector (Novagen, America) downstream of the 6-His tag and the constructed plasmid was verified using sequencing and restriction enzyme analysis. The construct was transformed into competent Escherichia coli and recombinant protein was expressed under the following conditions: the bacteria were cultured to reach an optical density (OD) of 0.6-0.8 at 600 nm (OD600) and were then induced with 1 mM/mL isopropyl-β-D- thiogalactopyranoside (IPTG) at 37°C for overnight. The cells were centrifuged at 12,000 rpm for 10 minutes at 4°C and were sonicated in lysis buffer (NaHPO4 100mM, NaCl 200 mM, Imidazole 5 mM, 1 mM PMSF with pH:8). Finally, the supernatant was evaluated for the presence of expressed protein by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and also was loaded over a Ni-nitrilotriacetic acid-agarose column (Qiagen, USA).
3.4. Western Blot Analysis of Recombinant Antigen
Extracted HCV core+1 protein was obtained as described above and subjected to SDS-PAGE and then, electro-transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore®). The PVDF membrane was incubated with 1X phosphate-buffered saline (PBS) and 0.5% skim milk overnight at 4°C and then, to determine the presence of core+1 Abs, the collected serum samples of subjects in both HCV core+1 positive/negative and control groups were used. After 2 hours, the paper was washed twice with PBS/Tween; then, the membranes were incubated with the peroxidase-conjugated secondary goat anti-human IgG (whole molecule)-peroxidase antibody (Sigma-Aldrich) for 2 hours. The membrane was subsequently washed twice more and protein bands were detected by the color reaction between diaminobenzidine (DAB) and horseradish peroxidase (Sigma, Germany). To determine the likelihood of possible core+1Ag cross-reactivity with the HCV-core protein, the specific antigenicity of the core+1 protein was assessed by Western blotting using both positive and negative anti-core+1 antibody HCV infected patients.
3.5. Detection of Anti-Core+1 and Anti-Core Antibodies by ELISA and Reproducibility Assay
Microplates were separately coated overnight with different concentration of 0.5, 1, and 2 μg/well of core and core+1 Ags in bicarbonate buffer pH 9.6 at 4°C. Afterwards, the wells were washed with PBS/Tween 20X and blocked with 1% gelatin in PBS for 2 hours at 37°C. The coated wells were washed 3 times with PBS/Tween 20X and serum samples as primary antibodies were added in dilution of 1:100, 1:500, 1:1000, 1:2000, and 1:4000 in duplicate for 2 hours at 37°C. Finally, the wells were washed and incubated with anti-human IgG (whole molecule)-peroxidase (Sigma-Aldrich) for 1 hour at 37°C followed by incubation with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate (Sigma, UK) and the reaction was stopped by H2SO4 2N and the OD was measured at 450 nm.
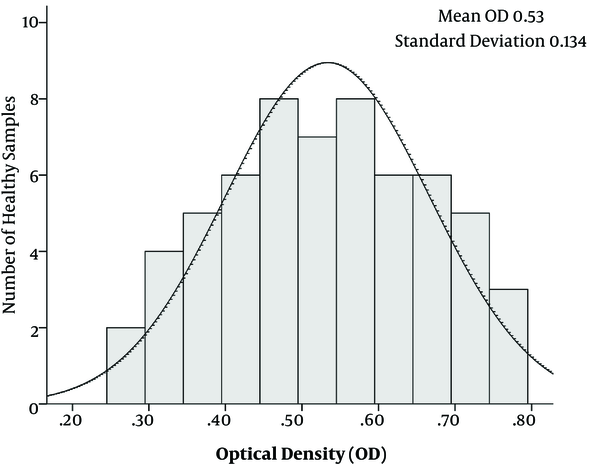
To avoid any nonspecific reactions, different set ups of ELISA tests were conducted; for instance, in different setups, one of the parameters including core+1 Ag, core Ag, patients’ sera as primary antibody and secondary antibody were deleted from the procedures. Moreover, different kinds of sera were incubated with each set of tests. All samples were tested in duplicates to determine the reproducibility of the developed ELISA. Fifteen positive samples and 25 negative samples were tested by 2 independent assays. The cut off value was calculated as the average OD of sera from the control group plus 3 standard deviations (SD) was determined as 0.93 (Figure 2).
The Value of Mean and Standard Deviation Was Calculated Based on OD of Samples from Healthy Controls
3.6. HCV Genotyping and Quantitation of HCV-RNA
Genotyping of HCV was performed using commercially available INNO-LiPA HCV II kit (Innogenetics, Belgium). The test was carried out according to the manufacture’s instruction. Viral RNA was extracted from 200 µL of serum using the Invisorb Spin Virus Mini Kit (Stratec Biomedical, Germany) according to the manufacturer’s protocol. Pretreatment HCV RNA was quantified by PCR (Roche COBAS Amplicor HCV monitor version 2.0, Roche Diagnostics, Mannheim, Germany).
3.7. Statistical Analysis
All statistical analyses were performed using the SPSS version 15 (SPSS Inc, USA). Variables were checked for normality by the Shapiro-Wilk test. Descriptive analysis including frequencies and cross tabs and paired sample t test, independent samples t test, and general linear model test were performed. Continuous variables were expressed as mean ± SD and categorical data were compared using χ2 test. Comparison of mean values between different groups was performed using the t test. A P value of less than 0.05 was considered statistically significant.
4. Results
The purified 24 kD core+1 and core Ags with the concentration of 1 μg/well were coated in ELISA plates. Moreover, the serum dilution of 1:500 was used as the primary set up to detect the individuals positive for anti-core+1 Abs. All the control groups were negative for anti-ARFP and anti-core antibodies. Furthermore, the result of Western blotting showed that only anti-core+1 antibody positive, but not anti-core positive sera reacted with core+1 protein that confirmed the specificity of anti-core+1 antibody and excluded the likelihood of the possible cross-reactivity of core+1/core-proteins (Figure 2).
Consecutive blood samples were obtained from the patients at the time point of before, during (12, 24, and 48 weeks), and 6 months after ETR. Responders were defined as patients with no detectable HCV RNA at the ETR and at least 6 months after ETR, while non-responders were defined as patients unclear for HCV RNA at the ETR. Seventy patients were included in the study. Among them, 34 patients (48.6%) did not respond to treatment (non-responders) and 36 patients (51.4%) cleared viremia for at least 6 months after ETR (responders). The mean age of the male subjects was lower than that of the female ones. The clinical characteristics of the patients are summarized in Table 1.
Demographic and Clinical Characteristics of the Study Participants
| Characteristic | Value | |
|---|---|---|
| Mean age ± SD (range) | 38.29 ± 12.11 (17 - 61) years | |
| Gender, n (%) | Male | 53 patients (75.7%) |
| Female | 17 patients (24.3%) | |
| HCV Genotype, n (%) | 1a | 70 patients (100%) |
| Groups | Responder | 36 patients (51.4%) |
| Non-responder | 34 patients (48.6%) | |
| Mean HCV load (copies/mL) ± SD | Responder | 2.7 × 10 6 ± 4.2 × 10 6 |
| Non-responder | 2.5 × 10 6 ± 3.5 × 10 6 | |
| Patients without documented HCV load | 10 patients | |
4.1. Prevalence of specific anti-core+1 Ab in responder and non-responder patients
The anti-core+1 Ab against the HCV core+1 protein with the concentration of 1 μg/well was detectable in the sera of patients infected with HCV with the dilution of 1:500. Samples were considered positive if the OD was above 0.93. Out of 70 patients infected with HCV, 53 (75.7%) were positive for anti-core+1 protein Abs before treatment including 27 (75%) responders and 26 (76.5%) non-responders. Seventeen patients (24.3%) were negative for anti-core+1 Abs before treatment, from which 9 patients (25%) were finally diagnosed as responders and 8 patients (23.5%) as non-responders. All the individuals in the control group were negative for anti-core+1 Ab.
4.2. Dynamics of the anti-core+1 Abs level during antiviral treatment and the role of anti-core+1 Abs in the outcome of treatment
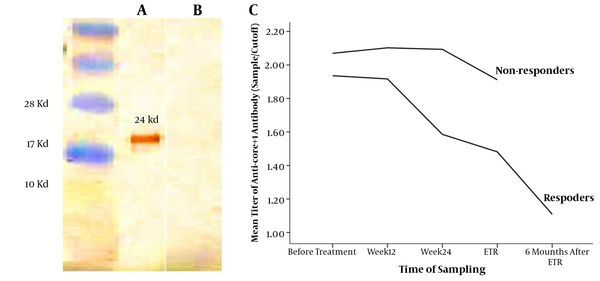
In order to investigate the effect of anti-core+1 Ab on the outcome of the treatment, patients infected with HCV were followed up for 48 weeks. Compared with the responders, the non-responder patients had a higher level of core+1 antibody titer against the core+1 protein in all of the research time point. As shown in Figure 3, the dynamic of anti-core+1 recombinant protein antibodies decreased during antiviral therapy in both groups of responders and non-responders, which was statistically significant just in responders (P < 0.0001, the paired t test). No patients with positive anti-core+1 Ab before treatment cleared the Ab at the end of treatment (week 48). Responders were followed-up for 6 months after termination of treatment and antibody titers were measured again at the week 72. Although the mean of the anticore+1 antibody of responders at the week 72 was significantly lower than that of the end of treatment, it was still detectable in 69.5% of responders.
HCV-Core + 1 Western Blot and Titer of Anti-Core + 1Abs
4.3. Correlation between the anti-core+1 Abs and other clinical parameters
There was no difference in the baseline viral load between the 2 groups of patients and it was not associated with the success of treatment (P = 0.506, the independent t test). Males had a higher pre-treatment viral load compared with females (P = 0.019, the independent t test), but baseline viral load was not correlated with anti-core+1 Ab titer. In the responders group, all patients (100%) showed a virological response, manifested as a negative PCR for HCV RNA. Overall, evidence showed that combination therapy affected the titer of anti-core+1 antibody in patients infected with HCV and the decrease in the titer of anti-core+1 Abs was less in the non-responder compared with responder patients.
5. Discussion
Core+1 protein is an HCV gene product expressed by transcriptional slippage resulting in the production of different ARFP isoforms with variable N-terminal regions (5, 14, 22-24). Although the function of the core+1 protein is not fully understood, there is evidence showing that core+1 protein may play a role in the entry mechanisms, morphogenesis, and regulation of cellular function, which are important in the viral life cycle (5). In the current study, the prevalence and titer of specific anti-core+1 Abs were determined in the serum of both responders and non-responders. The specific detection of purified core+1 protein by anti-core+1 polyclonal Abs was evaluated by ELISA and no cross-reactivity of core protein was observed. Western blotting was performed to demonstrate the specific antigenicity of the HCV core+1 protein compared with the HCV core protein. No cross-reactivity of anti-core Ab was observed with anti-core+1 Abs, despite the fact that both proteins possess the N-terminal domain of the core protein.
The fact that the most important antigenic domain of the HCV-core protein is located in amino acids 11 - 45 and the shared part of core+1 and core is the first 10 amino acids indicates the absence of cross-reactivity between the core and ARFP antigenic domains. The first 10 amino acids of the core+1 protein play a stabilizing role in the conformational structure of the core+1 protein, leading to better folding, rather than acting as an antigenic region (5). Therefore, this homology does not interfere with the anti-core+1 positive response (5); besides, the authors` previous study showed no cross-reactivity between the Abs against core+1, truncated core+1, and core proteins (10).
Previous reports indicated that the expression of core+1protein in patients infected with HCV was controversial (6). Therefore, the presence and titer of anti-core+1 Abs in sequential samples of 70 Iranian individuals infected with HCV treated with combination therapy were traced. Among these, 53 (75.7%) patients reacted positively with HCV core+1 recombinant protein. The seroprevalence levels of anti-core+1 Abs at baseline in responder and non-responder groups was 75% and 76.5%, respectively. This result was different from the ones reported by other groups (13, 17) that used recombinant core+1 antigen in ELISA, but were nearly similar to some other studies (5, 7).
In the current study, there was a significant change in the titer of anti-core+1 Abs during the combination therapy in responder patients compared to non-responder individuals, which was in accordance with some reports showing a link between the immune response to core+1 protein, manifested by the development of anti-core+1 Abs, and the outcome of combination therapy (5, 6, 11, 13, 17). In detail, Gao et al. (17) reported that from 72 patients infected with HCV receiving antiviral therapy, all of the patients with negative anti-core+1 Ab at the baseline and 70% of anti-core+1 Ab positive patients achieved SVR. Moreover, they showed that the therapy outcome was associated with a decline in titer in 2 of 5 cases by investigating the time-dependent changes in anti-core+1 Ab level. An earlier study from Iran also showed significant higher anti-core+1 antibody titer in patients resistant to HCV drug compared with patients showed SVR (11).
The current study findings revealed that the immune response to core+1 antigen was affected by the combination therapy. On the other hand, Cohen et al. (5), Bain et al., (6) and Karamitros et al., (13) in studies on of 27, 6, and 90 individuals infected with HCV respectively, found no changes in the titer of anti-core+1 Abs as well as no correlation between anti-core+1 Abs dynamics and the combination treatment outcome. It seems that the presence or absence of anti-core+1 antibody may not be a sensitive indicator for the prediction of the efficacy of combination therapy in patients infected with HCV, but the decreased titer of anti-core+1 Abs can be considered as an indicator to predict the outcome of therapy. Furthermore, it was found that the serum level of HCV core+1 antigen could not reflect the trend of HCV RNA level in patients with CHC undergoing combination therapy at baseline, that was compatible with some reports (13), and contrasted with the other reports (5, 17).
It could be due to the similar prevalence of anti-core+1 Abs in both responder and non-responder groups; therefore, it was thought that the higher viral load could not have a significant effect on the production of more core+1 protein in the ones that were positive for this antigen in both groups. Apart from methodology, parameters related to the selected study group such as race, HCV genotypes, and geographical regions as well as sample sizes might be the reasons that cause this difference. It seems that the mentioned factors might have some impacts on the HCV anti-core+1 Ab presence. Doing experiments on different groups infected with different genotypes might require a better understanding of how and why a patient generates different Ab responses against HCV core+1 protein.
To the authors best knowledge, the current study was the first study conducted to assess the effect of the titer of core+1 antibody on the outcome of treatment in the Iranian patients with CHC. The current study data were in agreement with some studies that showed the high F protein expression may be linked to the pathogenesis of the viral infection (1, 4). Here, it was suggested that only the lower titer of anti-core+1 antibody responses might be influenced by IFN combination therapy and associated with SVR in Iranian patients with HCV infection. On the other hand, there was a correlation between the immune response to core+1 protein and the response to combination therapy in patients infected with HCV as the cases with lower anti-core+1 activity were more likely to achieve SVR than other patients who were also positive for anti-core+1 antibodies. Therefore, the level of specific antibodies to core+1 protein may be considered as an alternative assessment of the therapeutic response in patients infected with HCV that can predict the efficacy of IFN therapy only in patients that develop an antibody response against HCV core+1 protein.
There were some studies showing that core+1 protein was associated with the outcomes of HCV infections, as this protein can suppress T-cell immune responses (19) and IFN production (21); therefore, complement available antiviral therapies, by blocking the inhibitory signaling pathways and some viral proteins such as core+1, may provide a specific new target to treat and prevent HCV infections. Due to small sample size of the current study, relevant studies are needed to determine whether the immune response of patients with CHC have a potential clinical value for diagnostic tests.
6. Conclusion
In conclusion, the current study showed that core+1 protein expressed during natural HCV infection and specific Ab could be detectable in most individuals chronically infected with HCV in both groups of responders and non-responders. Apparently, the titer of anti-core+1 Abs is related to response to treatment. Therefore, employment of this indicator can be useful to assess the therapeutic response in patients infected with HCV. In the context of HCV treatment, interventions to improve or modify treatment plan to enhance the outcome (here: SVR) with modification of dose and treatment duration of pegylated interferon-α (PegIFN-α) and ribavirin, or in combination with other new medicines to ensure that treatment will be successful, are suggested. The active or passive role of core+1 protein in the HCV life cycle and the significance of the effect of anti-core+1 Abs in the outcome of therapy still need to be elucidated.
Acknowledgements
References
-
1.
Ajorloo M, Bamdad T, Hashempour T, Alborzi AM, Mozhgani SH, Asadi R, et al. Detection of Specific Antibodies to HCV-ARF/CORE+1 Protein in Cirrhotic and Non-Cirrhotic Patients with Hepatitis C: A Possible Association with Progressive Fibrosis. Arch Iran Med. 2015;18(5):304-7. [PubMed ID: 25959912].
-
2.
Sarvari J, Mansouri M, Hashempoor T, Hosseini SY, Moattari A, Pirbonyeh N. Association of genotype and haplotype of IL-28B Gene with Hepatitis C infection outcome in iran: Spontaneous clearance versus chronic infection. Hepatit Month. 2017;17(5). e45745. https://doi.org/10.5812/hepatmon.45745.
-
3.
Alborzi A, Hashempour T, Moayedi J, Musavi Z, Pouladfar G, Merat S. Role of serum level and genetic variation of IL-28B in interferon responsiveness and advanced liver disease in chronic hepatitis C patients. Med Microbiol Immunol. 2017;206(2):165-74. [PubMed ID: 28214926]. https://doi.org/10.1007/s00430-017-0497-y.
-
4.
Dalagiorgou G, Vassilaki N, Foka P, Boumlic A, Kakkanas A, Kochlios E, et al. High levels of HCV core+1 antibodies in HCV patients with hepatocellular carcinoma. J Gen Virol. 2011;92(Pt 6):1343-51. [PubMed ID: 21307221]. https://doi.org/10.1099/vir.0.023010-0.
-
5.
Cohen M, Bachmatov L, Ben-Ari Z, Rotman Y, Tur-Kaspa R, Zemel R. Development of specific antibodies to an ARF protein in treated patients with chronic HCV infection. Dig Dis Sci. 2007;52(9):2427-32. [PubMed ID: 17436105]. https://doi.org/10.1007/s10620-006-9630-2.
-
6.
Bain C, Parroche P, Lavergne JP, Duverger B, Vieux C, Dubois V, et al. Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J Virol. 2004;78(19):10460-9. [PubMed ID: 15367612]. https://doi.org/10.1128/JVI.78.19.10460-10469.2004.
-
7.
Hashempour T, Ajorloo M, Bamdad T, Merat S, Zaer-Rezaee H, Fakharzadeh E, et al. Development of a recombinant based ELISA using specific antibodies to f protein in HCV chronically infected patients-a seroprevalence study. Iran J Virol. 2010;4(1):1-6. https://doi.org/10.21859/isv.4.1.1.
-
8.
Sarvari J, Mojtahedi Z, Taghavi SA, Kuramitsu Y, Shamsi Shahrabadi M, Ghaderi A, et al. Differentially Expressed Proteins in Chronic Active Hepatitis, Cirrhosis, and HCC Related to HCV Infection in Comparison With HBV Infection: A proteomics study. Hepat Mon. 2013;13(7):8351. [PubMed ID: 24066001]. https://doi.org/10.5812/hepatmon.8351.
-
9.
Gorzin AA, Ramsland PA, Tachedjian G, Gowans EJ. Identification of residues involved in NS2 homodimerization and elucidation of their impact on the HCV life cycle. J Viral Hepat. 2012;19(3):189-98. [PubMed ID: 22329373]. https://doi.org/10.1111/j.1365-2893.2011.01504.x.
-
10.
Hashempour T, Bamdad T, Bergamini A, Lavergne JP, Haj-Sheykholeslami A, Brakier-Gingras L. F protein increases CD4+CD25+ T cell population in patients with chronic hepatitis C. Pathog Dis. 2015;73(4). [PubMed ID: 25862675]. https://doi.org/10.1093/femspd/ftv022.
-
11.
Rahimi P, Vahabpour R, Fard FS, Motevalli F, Chahouki FF, Aghasadeghi MR, et al. Expression of HCV Alternative Reading Frame Protein (Core+1/F) in Baculovirus Expression System and its Evaluation for Assessment of Specific Anti-core+1 Antibody in Iranian HCV Infected Patients. Clin Lab. 2016;62(10):1919-26. [PubMed ID: 28164534]. https://doi.org/10.7754/Clin.Lab.2016.160205.
-
12.
Kassela K, Karakasiliotis I, Charpantidis S, Koskinas J, Mylopoulou T, Mimidis K, et al. High prevalence of antibodies to core+1/ARF protein in HCV-infected patients with advanced cirrhosis. J Gen Virol. 2017;98(7):1713-9. [PubMed ID: 28708052]. https://doi.org/10.1099/jgv.0.000851.
-
13.
Karamitros T, Kakkanas A, Katsoulidou A, Sypsa V, Dalagiorgou G, Mavromara P, et al. Detection of specific antibodies to HCV-ARF/CORE+1 protein in patients treated with pegylated interferon plus ribavirin. J Viral Hepat. 2012;19(3):182-8. [PubMed ID: 22329372]. https://doi.org/10.1111/j.1365-2893.2011.01502.x.
-
14.
Baril M, Brakier-Gingras L. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res. 2005;33(5):1474-86. [PubMed ID: 15755749]. https://doi.org/10.1093/nar/gki292.
-
15.
Shao SW, Wu WB, Bian ZQ, Yu JG, Zhao P, Zhao LJ, et al. Hepatitis C virus F protein inhibits cell apoptosis by activation of intracellular NF-kappaB pathway. Hepatol Res. 2009;39(3):282-9. [PubMed ID: 19054148]. https://doi.org/10.1111/j.1872-034X.2008.00452.x.
-
16.
Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25(1):105-17. [PubMed ID: 15732002]. https://doi.org/10.1055/s-2005-864786.
-
17.
Gao DY, Zhang XX, Hou G, Jin GD, Deng Q, Kong XF, et al. Assessment of specific antibodies to F protein in serum samples from Chinese hepatitis C patients treated with interferon plus ribavarin. J Clin Microbiol. 2008;46(11):3746-51. [PubMed ID: 18832124]. https://doi.org/10.1128/JCM.00612-08.
-
18.
Xiao W, Jiang LF, Deng XZ, Zhu DY, Pei JP, Xu ML, et al. PD-1/PD-L1 signal pathway participates in HCV F protein-induced T cell dysfunction in chronic HCV infection. Immunol Res. 2016;64(2):412-23. [PubMed ID: 26286967]. https://doi.org/10.1007/s12026-015-8680-y.
-
19.
Pei JP, Jiang LF, Ji XW, Xiao W, Deng XZ, Zhou ZX, et al. The relevance of Tim-3 polymorphisms and F protein to the outcomes of HCV infection. Eur J Clin Microbiol Infect Dis. 2016;35(8):1377-86. https://doi.org/10.1007/s10096-016-2676-y.
-
20.
Kotta-Loizou I, Karakasiliotis I, Vassilaki N, Sakellariou P, Bartenschlager R, Mavromara P. Expression of the novel hepatitis C virus core+1/ARF protein in the context of JFH1-based replicons. J Virol. 2015;89(9):5164-70. [PubMed ID: 25694591]. https://doi.org/10.1128/JVI.02351-14.
-
21.
Park SB, Seronello S, Mayer W, Ojcius DM. Hepatitis C Virus Frameshift/Alternate Reading Frame Protein Suppresses Interferon Responses Mediated by Pattern Recognition Receptor Retinoic-Acid-Inducible Gene-I. PLoS One. 2016;11(7). e0158419. [PubMed ID: 27404108]. https://doi.org/10.1371/journal.pone.0158419.
-
22.
Vassilaki N, Mavromara P. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J Biol Chem. 2003;278(42):40503-13. [PubMed ID: 12874283]. https://doi.org/10.1074/jbc.M305504200.
-
23.
Boulant S, Becchi M, Penin F, Lavergne JP. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem. 2003;278(46):45785-92. [PubMed ID: 12952944]. https://doi.org/10.1074/jbc.M307174200.
-
24.
Qureshi H, Qazi R, Hamid S, Qureshi SA. Identification of immunogenic regions within the alternative reading frame protein of hepatitis C virus (genotype 3). Eur J Clin Microbiol Infect Dis. 2011;30(9):1075-83. [PubMed ID: 21318731]. https://doi.org/10.1007/s10096-011-1194-1.