Abstract
Background:
Campylobacteriosis is a zoonotic infectious disease caused by Campylobacter jejuni and C. coli. The cadF gene is considered as a genus-specific gene while other genes are mainly used for discrimination at the species level.Objectives:
This study aimed to analyze the cadF gene and to develop a duplex PCR assay for simultaneous detection of C. coli and C. jejuni, the two commonly encountered species.Materials and Methods:
In silico analysis of the cadF gene was carried out by several software and available online tools. A duplex PCR optimized with specific primers was used for detection and differentiation of both species. To evaluate specificity and sensitivity of the test, a panel of different Campylobacter spp. together with several intestinal bacterial pathogens was tested. The limit of detection (LOD) of method was determined using serial dilutions of standard genomes.Results:
The analysis of the full size cadF gene indicated variations in this gene, which can be used to differentiate C. jejuni and C. coli. The duplex PCR designed in this study showed that it could simultaneously detect and differentiate both C. jejuni and C. coli with product sizes of 737 bp and 461 bp, respectively. This assay, with 100% specificity and sensitivity, had a limit of detection (LOD) of about 14 and 0.7 µg/mL for C. jejuni and C. coli, respectively.Conclusions:
In silico analysis of the cadF full-gene showed variations between the two species that can be used as a molecular target for differentiating C. jejuni and C. coli in a single-step duplex-PCR assay with high specificity and sensitivity.Keywords
1. Background
Campylobacter enteritis is one of the most frequent food-borne infections worldwide (1). Thermophilic C. jejuni and C. coli have been recognized as the most common causes of bacterial diarrhea in humans, especially among children less than five years of age and young adults (2). Although, poultry and poultry products are important sources of Campylobacteriosis, yet the organism can be transmitted to humans via contact with other warm-blooded animals such as cattle, pigs, sheep, ostriches, shellfish, and pets (3, 4).
The symptoms of campylobacteriosis can vary from mild to severe complications, including abdominal pain, fever, myalgia and watery or bloody diarrhea. Although, in most cases the illness is self-limited and rarely fatal yet post-infectious acute immune-mediated neurologic complications such as Guillain-Barre syndrome and Miller Fisher syndrome can occur, which are the consequence of molecular mimicry between lipooligosaccharides (LOS) of bacterial cell wall and gangliosides in peripheral nerves of humans (5, 6). These complications can be prevented or lowered with rapid and accurate detection of etiological agents of the disease. Diagnosis of campylobacteriosis is performed through microbiological, molecular and serological tests. Culture is the gold standard of diagnosis of C. coli and C. jejuni; however, the culture conditions for detection of these fastidious bacteria are complicated and time consuming, which in some cases make the recovery of bacteria unsuccessful. Moreover, the emergence of viable but non-culturable (VBNC) phenotypes should not be ignored.
Differentiation of the two species is only performed through hippurate hydrolysis biochemical test or molecular-based detections (7-10). In molecular methods different genetic targets have been used for the detection Campylobacter species (e.g. asp, hipO, ceuE,cadF, 16SrRNA, 23S rRNA and cdt, fur, glyA, cdtABC, ceuB–E and fliY) (9). Among them, the cadF gene encodes a fibronectin-binding protein that promotes bacteria-host cell interaction and has been described as a conserve and genus-specific gene. In most studies a fragment from this gene with a length of 400 bp is used for identification of Campylobacter spp. at the genus level (1, 11-15). There is no documented bioinformatics study on the cadF gene full-sequence analysis in C. jejuni and C. coli.
2. Objectives
The aim of this study was to analyze the cadF gene and to develop and evaluate a single-step duplex polymerase chain reaction (PCR) assay for simultaneous detection of C. coli and C. jejuni, the two commonly encountered species in human Campylobacteriosis.
3. Materials and Methods
3.1. Alignment of cadF Sequences From GenBank
The cadF sequences from the complete genome of C. jejuni and C. coli were acquired from NCBI GenBank (http://www.ncbi.nlm.nih.gov/) (Table 1). Multiple alignments were performed using the CLC sequence viewer 7.6 software (CLC bio, Aarhus, Denmark).
CampylobactercadF Sequences Used in This Study
| Definition | Accession No. |
|---|---|
| Campylobacter jejuni subsp. jejuni strain MTVDSCj20, complete genome | CP008787.1 |
| Campylobacter jejuni subsp. jejuni 00-2538, complete genome | CP006707.2 |
| Campylobacter jejuni subsp. jejuni 00-2544, complete genome | CP006709.2 |
| Campylobacter jejuni subsp. jejuni PT14, complete genome | NC_018709.2 |
| Campylobacter jejuni subsp. jejuni NCTC 11168 complete genome | AL111168.1 |
| Campylobacter jejuni RM1221, complete genome | CP000025.1 |
| Campylobacter jejuni subsp. jejuni 81116, complete genome | CP000814.1 |
| Campylobacter coli RM2228 cont193, whole genome shotgun sequence | AAFL01000010.1 |
| Campylobacter coli RM1875, complete genome | CP007183.1 |
| Campylobacter coli 15-537360, complete genome | CP006702.1 |
| Campylobacter coli RM5611, complete genome | CP007179.1 |
| Campylobacter coli CVM N29710, complete genome | CP004066.1 |
| Campylobacter coli RM4661, complete genome | CP007181.1 |
| Campylobacter coli JV20 contig00034, whole genome shotgun sequence | AEER01000022.1 |
| Campylobacter coli JV20 genomic scaffold SCAFFOLD1, whole genome shotgun sequence | GL405235.1 |
3.2. In Silico Analysis of the cadF Gene
The conserved internal fragment (400 bp) of the cadF gene, reported by Konkel et al. (15) as a specific gene for detection of Campylobacter spp., was used as a reference sequence in this study. This fragment and other selected sequences from the full gene of cadF were subjected to in silico analysis with the online NEB cutter program (http://tools.neb.com/NEBcutter) to compare and select a proper restriction endonuclease for discriminating between C. jejuni and C. coli using analysis of enzymatic digestion pattern.
3.3. Designing a Duplex Polymerase Chain Reaction Assay for Specific Detection of Campylobacter jejuni and C. coli
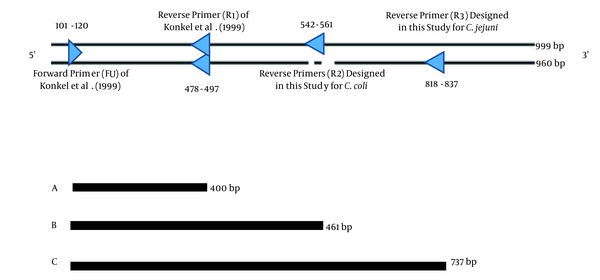
The entire cadF sequence obtained from GenBank was robustly examined for the presence of intra-species conserved regions, which could differentiate inter-species. Universal forward primer, FU, (position 101 - 120) and reverse primer, R1, (position 478 - 497) were selected for the cadF gene, and were previously described by Konkel et al. (15). Other reverse primers, R2 (position 542 - 561) and R3 (position 818 - 837), were designed in this study using the Genrunner and CLC sequence viewer software (Table 2). Analysis of the designed primers was performed by the Primer-BLAST on NCBI (http://www. ncbi.nlm.nih.gov/). Schematic representation of the PCR amplification of fragments related to C. jejuni and C. coli in duplex PCR is shown in Figure 1. Oligonucleotide primers were synthesized by TAG Copenhagen (Denmark).
Position of Primers for Developing the Duplex Polymerase Chain Reaction Assay
The duplex-PCR was carried out in a 25-μL reaction mixture, containing 10 ng of DNA template extracted by the boiling method, 2.5 μL PCR buffer 10X, 200 μM dNTP, 5 mM MgCl2, 0.1 μM of each primer, 1 unit of Taq DNA polymerase, and sterile deionized water (12, 14). Amplification conditions were 95°C for three minutes (one cycle), then denaturation at 94°C for 30 seconds, annealing at 43°C for 30 seconds and extension at 72°C for 30 seconds for 32 cycles in a thermocycler (Eppendorf, Hamburg, Germany). Finally, an additional extension step (five minutes, 72°C) was carried out.
3.4. Limit of Detection, Sensitivity and Specificity of the Duplex Polymerase Chain Reaction
Limit of detection (LOD) of the amplification assay was evaluated using serial 10-fold dilutions of genomes with initial concentrations of 140 (C. jejuni) and 7 (C. coli) µg/mL. A total of 20 clinical and environmental Campylobacter isolates were examined for further evaluation of the sensitivity. Specificity of the test was evaluated using genomic DNA from standard and isolated clinical strains of other enteric non-Campylobacter bacterial pathogens (Table 3). Sensitivity and specificity were calculated according to the following Equations (16):
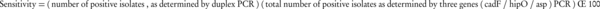
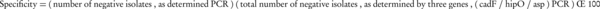
List of Bacteria Used for the Determination of Specificity and Sensitivity of cadF-Targeted Species-Specific Duplex Polymerase Chain Reactiona
| Organism Name | Strain Name | Amplification with Newly Designed Primers |
|---|---|---|
| Shigella sonnei | ATCC 25931 | negative |
| Shigella flexneri | ATCC 12022 | negative |
| Shigella boydii | ATCC 8700 | negative |
| Shigella dysenteriae | ATCC 13313 | negative |
| Aeromonas hydrophila | ATCC 7966 | negative |
| Enterobacter aerogenes | ATCC 13048 | negative |
| Vibrio cholerae | ATCC 39315 | negative |
| Enteropathogenic Escherichia coli | ATCC 43887 | negative |
| Escherichia coli O157:H7 | ATCC 35150 | negative |
| Enteroinvasive Escherichia coli | ATCC 43893 | negative |
| Enteroaggregative Escherichia coli | ATCC 33780 | negative |
| Enterotoxigenic Escherichia coli | ATCC 35401 | negative |
| Salmonella typhimurium | ATCC 29946 | negative |
| Salmonella typhi | ATCC 19430 | negative |
| Campylobacter jejuni | ATCC 29428 | positive |
| Campylobacter coli | ATCC 43478 | positive |
| Campylobacter coli | Isolate 1 | positive |
| Campylobacter coli | Isolate 2 | positive |
| Campylobacter coli | Isolate 3 | positive |
| Campylobacter coli | Isolate 4 | positive |
| Campylobacter coli | Isolate 5 | positive |
| Campylobacter coli | Isolate 6 | positive |
| Campylobacter coli | Isolate 7 | positive |
| Campylobacter coli | Isolate 8 | positive |
| Campylobacter coli | Isolate 9 | positive |
| Campylobacter jejuni | Isolate 1 | positive |
| Campylobacter jejuni | Isolate 2 | positive |
| Campylobacter jejuni | Isolate 3 | positive |
| Campylobacter jejuni | Isolate 4 | positive |
| Campylobacter jejuni | Isolate 5 | positive |
| Campylobacter jejuni | Isolate 6 | positive |
| Campylobacter jejuni | Isolate 7 | positive |
| Campylobacter jejuni | Isolate 8 | positive |
| Campylobacter jejuni | Isolate 9 | positive |
| Campylobacter jejuni | Isolate 10 | positive |
| Campylobacter jejuni | Isolate 11 | positive |
4. Results
4.1. In Silico Analysis of the cadF Gene
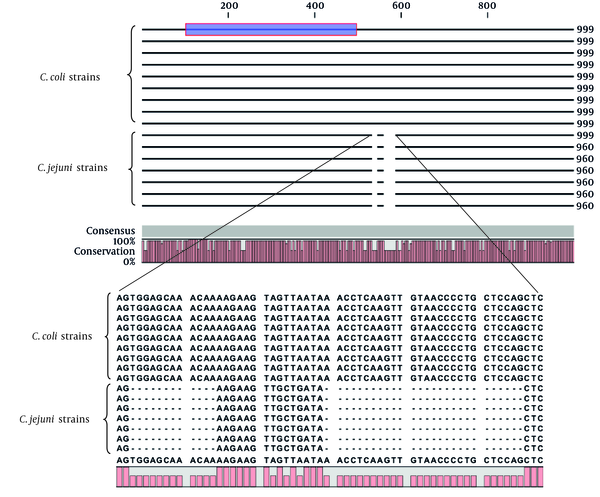
The length of the cadF sequences extracted from complete genome of C. jejuni and C. coli was 960 (with C + G 31.8% and A + T 68.2%) and 999 bp (with C + G 34% and A + T 66%), respectively. The cadF gene in both species was located after the rpsI gene, which coded for a 30S ribosomal protein. Although there were some nucleotide variations along the sequence of cadF between the two species, yet the main difference was related to the 39-bp deletion in the positions of 533 - 544 and 560 - 586 of C. jejuni (Figure 2). The results of the BLAST analysis of cadF gene showed an average sequence identity of 98.5% and 94% among C. jejuni and C. coli strains, respectively. The identity between the two species was also estimated as 88%, approximately (Table 4).
Analysis and Multiple Alignment of the cadF Complete Gene of Campylobacter jejuni (960 bp) and C. coli (999 bp) by the CLC Software
The Distribution of Nucleotides and the Percentage Identity of the cadF Gene
The analysis of the cadF gene 400-bp product sequence (Figure 2), introduced by Konkel et al. via the NEBcutter online web site, indicated that this part of the gene is conserved and there are no commercially proper restriction enzymes to produce fragments with good intervals to differentiate between C. jejuni and C. coli (15) The selected segments of the cadF gene in this study were theoretically appropriate for enzymatic digestions and the produced fragments could be used for species differentiation.
4.2. Design and Evaluation of a Species-Specific Duplex Polymerase Chain Reaction Assay
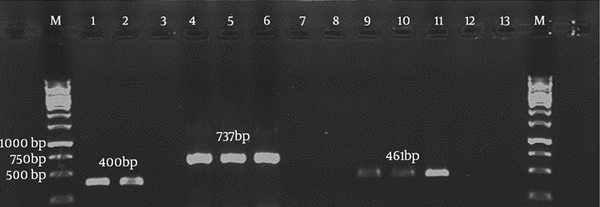
Duplex PCR showed that it could simultaneously detect both C. jejuni and C. coli with product sizes of 737 bp and 461 bp, respectively (Figure 3). Specificity and sensitivity of the duplex PCR assay was determined to be 100% with exclusive amplification for C. jejuni and C. coli, while this result was negative for other non-Campylobacter enteric bacterial species. The assay showed limit of detection (LOD) of 14 and 0.7 µg/mL (approximately equal to 7 × 109 and 3 × 108 copy number) for C. jejuni and C. coli, respectively.
Agarose Gel Electrophoresis of the Duplex Polymerase Chain Reaction Assay with Specific Primers
5. Discussion
Campylobacter jejuni and C. coli are now recognized as important causes of acute bacterial diarrhea in most countries. The isolation and discrimination of C. jejuni and C. coli by biochemical tests at the species level is limited and laborious, thus there is a crucial need to develop a sensitive, validated and rapid DNA-based method for detection of Campylobacter at the species level (10, 17). In some studies, multiple genes have been used for distinguishing C. jejuni and C. coli. Al Amri et al. (2007) developed a multiplex PCR assay using the combination of a genus-specific virulence gene (cadF) together with hippuricase and aspartokinase genes (asp) for species-specific identification of C. jejuni and C. coli, respectively (11). In a study by Cloak and Fratamico (2002), a multiplex PCR was designed for differentiation of C. jejuni and C. coli by means of cadF and ceuE genes. In another work by Adzitey and Corry (2011), lpxA, hipO and glyA genes were used for differentiating C. jejuni and C. coli species. The study of Nayak et al. (2005) was also designed with cadF, ceuE and oxidoreductase subunit genes as fragments of 400-bp conserved region in Campylobacter spp. 894-bp specific for C. coli and 160-bp specific for C. jejuni, respectively (14, 18, 19).
Our duplex PCR method was developed only with the cadF gene and the specificity and sensitivity of novel reverse primers (R2 and R3) in association with a previously described forward primer (FU) was studied. The PCR assay designed in this work showed 100% sensitivity and specificity while no amplification product was seen for the genomic DNA from non-Campylobacter enteric bacteria. One applicable advantage of this newly designed duplex PCR assay is that the amplified products are of different sizes, which can be concurrently visualized on agarose gel without the need to duplicate the reaction or further electrophoresis and sequencing. In a similar study, Klena et al. used divergence and conservation regions of lpxA to develop a robust PCR assay. They differentiated C. coli, C. jejuni, C. lari and C. upsaliensis using multiplex PCR with the lipid A gene lpxA, encoding a UDP-N- acetyl glucosamine acyl transferase. Another work similar to our research was the study of Gonzalez et al. They discriminated C. jejuni and C. coli by using ceuE gene diversity (approximately 13%) between two species (20, 21).
The lowest concentration of genomic DNA for detection of C. jejuni and C. coli was 14 and 0.7 µg/mL (approximately equal to 7 × 109 and 3 × 108 copy number), respectively. These LODs are almost comparable with the study conducted by Wisessombat et al., in which the sensitivity of the multiplex PCR for the detection of Campylobacter spp. was 2 × 105 CFU/PCR (22). Another study indicated that the colony multiplex PCR sensitivity range for C. jejuni and C. coli was 108 to 1013 and 106 to 1013 CFU/mL, respectively (23).
The bioinformatics data analysis of the 400-bp internal section of the cadF introduced by Konkel et al. which has been used by many investigators for genus-specific detection of Campylobacter spp. showed that this fragment is highly conserved among C. jejuni and C. coli strains and is significantly validated for the identification of both species. It seems that there is a concomitant general misjudged belief that the cadF full-gene is genus-specific. Our analysis of total cadF sequence revealed that other than single-nucleotide variations between two bacteria, an approximately 4% deletion has occurred in the cadF sequence of C. jejuni compared with C. coli, which could be useful for our work. The intra-species identity level among C. jejuni and C. coli strains was about 98.5% and 94%, respectively. The identity level was approximately 88% between the two species. These results were similar to the report of Konkel et al. with 87% identity between C. jejuni and C. coli and 98.6% among C. jejuni strains, individually (15).
There are several articles about the PCR-RFLP method for the differentiation of Campylobacter spp. using genes other than cadF (24, 25). Although the restriction pattern of enzymatic digestion of the 400-bp fragment introduced by Konkel et al. is not suitable for separation of the two species, yet the enzymatic digestion of the full-length gene may be useful for differentiation and clinical diagnosis of C. jejuni and C. coli (15) The cadF full-gene has some variations in its sequence and length between species, which can be beneficial for developing a duplex PCR. The designed PCR assay in this study is highly sensitive and specific and provides an accurate, inexpensive, sensitive and specific tool for rapid and simultaneous detection and differentiation of C. coli and C. jejuni in clinical settings.
References
-
1.
Randall L, Lemma F, Rodgers J, Vidal A, Clifton-Hadley F. Development and evaluation of internal amplification controls for use in a real-time duplex PCR assay for detection of Campylobacter coli and Campylobacter jejuni. J Med Microbiol. 2010;59(Pt 2):172-8. [PubMed ID: 19833779]. https://doi.org/10.1099/jmm.0.014415-0.
-
2.
Galanis E. Campylobacter and bacterial gastroenteritis. CMAJ. 2007;177(6):570-1. [PubMed ID: 17846438]. https://doi.org/10.1503/cmaj.070660.
-
3.
Sarkar SR, Hossain MA, Paul SK, Ray NC, Sultana S, Rahman MM, et al. Campylobacteriosis - an overview. Mymensingh Med J. 2014;23(1):173-80. [PubMed ID: 24584395].
-
4.
World Health Organization. Campylobacter Fact Sheet N255. Geneva: WHO; October 2011. Available from: http://www.who.int/mediacentre/factsheets/fs255/en/.
-
5.
Haag LM, Fischer A, Otto B, Grundmann U, Kuhl AA, Gobel UB, et al. Campylobacter jejuni infection of infant mice: acute enterocolitis is followed by asymptomatic intestinal and extra-intestinal immune responses. Eur J Microbiol Immunol (Bp). 2012;2(1):2-11. [PubMed ID: 24611115]. https://doi.org/10.1556/EuJMI.2.2012.1.2.
-
6.
Godschalk PC, Kuijf ML, Li J, St Michael F, Ang CW, Jacobs BC, et al. Structural characterization of Campylobacter jejuni lipooligosaccharide outer cores associated with Guillain-Barre and Miller Fisher syndromes. Infect Immun. 2007;75(3):1245-54. [PubMed ID: 17261613]. https://doi.org/10.1128/IAI.00872-06.
-
7.
Williams LK, McMeechan A, Baalham T, Ward L, Humphrey TJ, Jorgensen F. Detection of Campylobacter jejuni and Campylobacter coli from broiler chicken-related samples using BAX PCR and conventional International Organization for Standardization culture. J Food Prot. 2008;71(4):835-8. [PubMed ID: 18468043].
-
8.
Jackson DN, Davis B, Tirado SM, Duggal M, van Frankenhuyzen JK, Deaville D, et al. Survival mechanisms and culturability of Campylobacter jejuni under stress conditions. Antonie Van Leeuwenhoek. 2009;96(4):377-94. [PubMed ID: 19774483]. https://doi.org/10.1007/s10482-009-9378-8.
-
9.
Persson S, Olsen KE. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol. 2005;54(Pt 11):1043-7. [PubMed ID: 16192435]. https://doi.org/10.1099/jmm.0.46203-0.
-
10.
Kulkarni SP, Lever S, Logan JM, Lawson AJ, Stanley J, Shafi MS. Detection of campylobacter species: a comparison of culture and polymerase chain reaction based methods. J Clin Pathol. 2002;55(10):749-53. [PubMed ID: 12354800].
-
11.
Al Amri A, Senok AC, Ismaeel AY, Al-Mahmeed AE, Botta GA. Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J Med Microbiol. 2007;56(Pt 10):1350-5. [PubMed ID: 17893173]. https://doi.org/10.1099/jmm.0.47220-0.
-
12.
Ghorbanalizadgan M, Bakhshi B, Kazemnejad Lili A, Najar-Peerayeh S, Nikmanesh B. A molecular survey of Campylobacter jejuni and Campylobacter coli virulence and diversity. Iran Biomed J. 2014;18(3):158-64. [PubMed ID: 24842142].
-
13.
Patrone V, Campana R, Vallorani L, Dominici S, Federici S, Casadei L, et al. CadF expression in Campylobacter jejuni strains incubated under low-temperature water microcosm conditions which induce the viable but non-culturable (VBNC) state. Antonie Van Leeuwenhoek. 2013;103(5):979-88. [PubMed ID: 23314927]. https://doi.org/10.1007/s10482-013-9877-5.
-
14.
Nayak R, Stewart TM, Nawaz MS. PCR identification of Campylobacter coli and Campylobacter jejuni by partial sequencing of virulence genes. Mol Cell Probes. 2005;19(3):187-93. [PubMed ID: 15797819]. https://doi.org/10.1016/j.mcp.2004.11.005.
-
15.
Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol. 1999;37(3):510-7. [PubMed ID: 9986804].
-
16.
Roozbehani AD, Bakhshi B, Pourshafie MR, Katouli M. A rapid and reliable species-specific identification of clinical and environmental isolates of Vibrio cholerae using a three-test procedure and recA polymerase chain reaction. Indian J Med Microbiol. 2012;30(1):39-43. [PubMed ID: 22361759]. https://doi.org/10.4103/0255-0857.93027.
-
17.
Keramas G, Bang DD, Lund M, Madsen M, Bunkenborg H, Telleman P, et al. Use of culture, PCR analysis, and DNA microarrays for detection of Campylobacter jejuni and Campylobacter coli from chicken feces. J Clin Microbiol. 2004;42(9):3985-91. [PubMed ID: 15364980]. https://doi.org/10.1128/JCM.42.9.3985-3991.2004.
-
18.
Cloak OM, Fratamico PM. A multiplex polymerase chain reaction for the differentiation of Campylobacter jejuni and Campylobacter coli from a swine processing facility and characterization of isolates by pulsed-field gel electrophoresis and antibiotic resistance profiles. J Food Prot. 2002;65(2):266-73. [PubMed ID: 11848556].
-
19.
Adzitey F, Corry J. A Comparison between Hippurate Hydrolysis and Multiplex PCR for Differentiating Campylobacter coli and Campylobacter jejuni. Trop Life Sci Res. 2011;22(1):91-8. [PubMed ID: 24575212].
-
20.
Klena JD, Parker CT, Knibb K, Ibbitt JC, Devane PM, Horn ST, et al. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J Clin Microbiol. 2004;42(12):5549-57. [PubMed ID: 15583280]. https://doi.org/10.1128/JCM.42.12.5549-5557.2004.
-
21.
Gonzalez I, Grant KA, Richardson PT, Park SF, Collins MD. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol. 1997;35(3):759-63. [PubMed ID: 9041429].
-
22.
Wisessombat S, Inthagard J, Kittiniyom K, Srimanote P, Wonglumsom W, Voravuthikunchai SP. Multiplex PCR for direct identification of thermophilic campylobacter, C. jejuni, C. coli, C. lari and C. upsaliensis and simultaneous detection of CDTB gene. J Rapid Methods Autom Microbiol. 2009;17(4):438-54.
-
23.
Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol. 2002;40(12):4744-7. [PubMed ID: 12454184].
-
24.
Kashoma IP, Kumar A, Sanad YM, Gebreyes W, Kazwala RR, Garabed R, et al. Phenotypic and genotypic diversity of thermophilic Campylobacter spp. in commercial turkey flocks: a longitudinal study. Foodborne Pathog Dis. 2014;11(11):850-60. [PubMed ID: 25184688]. https://doi.org/10.1089/fpd.2014.1794.
-
25.
Kamei K, Asakura M, Somroop S, Hatanaka N, Hinenoya A, Nagita A, et al. A PCR-RFLP assay for the detection and differentiation of Campylobacter jejuni, C. coli, C. fetus, C. hyointestinalis, C. lari, C. helveticus and C. upsaliensis. J Med Microbiol. 2014;63(Pt 5):659-66. [PubMed ID: 24568882]. https://doi.org/10.1099/jmm.0.071498-0.