Abstract
Background:
Exploring the rate of naturally occurring NS3 protease mutants in HCV infected population is influential in the future therapeutic approaches.Objectives:
This study explored naturally occurring resistant mutations to protease inhibitors in a pilot study.Patients and Methods:
We analyzed NS3 gene sequences in 7 HCV infected patients, referred to the central liver center, south of Iran. The protease domain was amplified by PCR followed by product extraction. Amplified NS3 genes were cloned by TA/cloning system. For each patient, clonal-sequencing was performed to improve mutation detection sensitivity. Then, the obtained sequences were compared with the reference sequences and final phylogenic tree was constructed. Afterwards, the sequences were studied to investigate point mutations.Results:
Phylogenetic analysis between reference and amplified sequences demonstrated high similarity of all sequences with genotype 1. Interestingly, crucial protease resistant mutations were detected in V36 and R155 positions in one patient’s sequence. Checking different clones of this patient confirmed V36L, as the dominant mutation while R155K was detected only in a few cases.Conclusions:
As revealed, naturally occurring resistant mutations, especially R155K in protease sequence were identified in 1 out of the 7 patients, so the rate of such mutations is estimated to be high. It seems that checking HCV patients before protease inhibitor treatment are necessary in the region.Keywords
1. Background
Hepatitis C virus (HCV) infection remains a serious health concern, with an estimated prevalence of more than 170 million cases worldwide. The prevalence of this infection is less than 1% in Iranian blood donors although the total rate of infection is rising in developing countries (1). Despite the standard therapy, including recombinant interferon and ribavirin, significant side effects and failed responses in a large proportion of patients, particularly those infected with genotype 1 virus, still remain a challenge (2).
Recently, to overcome these shortcomings, different generations of HCV antiviral drugs have been produced, among them protease inhibitors (PIs) have shown promising results in late-stages of clinical trials (3-5). PIs drugs inhibit NS3 protease activity by attaching either to activation site or surrounding motifs (6). In both cases, PIs block enzyme activity and due to crucial enzyme function, they limit viral replication both in vitro and in vivo (7, 8). NS3 protease activity is related to 189 amino acids from N-terminal part of protein. HCV like other RNA viruses, due to its nature of RNA-dependent RNA polymerase (RDRP), is prone to point mutation even in the absence of environmental bottlenecks (9). Sequence diversity in HCV is responsible for natural drug resistance against upcoming therapy like PIs and remains one of the major concerns for physicians (3, 10).
Natural mutations in NS3 protease region appear to be due the nature of polymerase enzyme and have been evaluated in some previous studies (11, 12). Availability of these drugs in developing countries (before their widespread use) makes the screening of the specific natural mutations in NS3 protease warranted based on their cost–effectiveness and susceptibility (7, 13). Effective PIs resistant mutations have been characterized in different studies and their impacts on treatment outcomes have also been delineated recently (7, 13, 14).
To our knowledge, limited data about the natural resistance to PIs have been published from the Middle East. In this preliminary study, we attempted to find the first HCV infected patient with natural PIs resistance before embarking on a wider project. To this effect, a small group of PIs native patients referred to a Liver Center, south of Iran, were screened for PIs resistant mutations. Furthermore, to enhance the sensitivity of detection, clonal-sequencing approach was employed instead of crude sequencing.
2. Objectives
Our aim was to conduct a preliminary study to estimate the rate of NS3 PIs resistance among a small population of HCV patients.
3. Patients and Methods
3.1. Patients
During autumn 2013, a total of 10 chronic HCV patients referred to the liver clinic of Shahid Motahari hospital (Shiraz, Iran), were randomly selected and enrolled in this study. They were between 18 and 50 years of age (mean age 38 ± 6 y, 7 men and 3 women). Besides, informed consents were obtained from all and recommended guidelines from local ethics committee, Shiraz university of medical sciences were fully considered. All patients had received a course of standard peg-interferon/ribavirin therapy but none of them consumed PIs drugs at the sampling time. Also, they were confirmed as negative cases of HIV and HBV infection. Blood samples were taken in EDTA tubes and plasma parts were collected in sterile condition, then, they were stored at -70°C until further tests.
3.2. Primer Design
At first, the HCV genotypes and subtypes (those with genotype 1a/or 1b) full length sequences, were retrieved from NCBI (National Center for biotechnology Information) and LOS Alamos database. Then, multiple alignments were performed by Bioedit and Clustal X softwares. The primer sequences covering 203 amino acids from protease region of NS3, were designed by Oligo7 and primer3 softwares so that they can detect the sequences associated with genotype 1a and 1b. The primers are listed in Table 1.
Designed Primers and Their Positions According to H77 Virus Reference Sequence
| Primer Name | Sequence 5’ - 3’ | Position | Function |
|---|---|---|---|
| For-1 (Common) | CACATGGAGAYCAAGVTCATCAC | 3273 - 3295 | First amplification primer (common for genotypes) |
| Rev-1 (Common) | RGCCGGSACCTTDGTGCT | 4067 - 4050 | First amplification primer (common for genotypes) |
| F2G1A | GCGTGCGGTGACATCATC | 3315 - 3332 | Second amplification primer (specialized for genotype 1a) |
| F2G1B | GCGTGTGGGGACATCATC | 3315 - 3332 | Second amplification primer (specialized for genotype 1b) |
| Rev-2 (Common) | CTTRGTGCTCTTRCCGCTGC | 4058 - 4039 | Second amplification primer (common for genotypes) |
| NCR1 | CCCCTGTGAGGAACTACTGTC | 42 - 62 | First round detection of HCV |
| NCR2 | TGCACGGTCTACGAGACCTC | 339 - 320 | First round detection of HCV |
| NCR3 | CACGCAGAAAGCGTCTAGCCATG | 65 - 87 | Nested round detection of HCV |
| NCR4 | GCGTGCGGTGACATCATC | 310 - 289 | Nested round detection of HCV |
Besides, to confirm the presence of virus genome inside the samples before starting the protocol, an in-house nested PCR assay was performed by specific primers of 5/noncoding region, NCR. These primers were designed to amplify early 5/NCR region of all genotypes. Sequences of primers for this purpose are shown in Table 1 and nominated as NCR1 and NCR2 for the first round and NCR3 and NCR4 for nested round amplification of target.
3.3. Amplification of NS3 Protease Region
Viral RNA was extracted from the serum samples using RNA extraction kit (Invitek, Germany), according to the respective instructions. Complementary DNA (cDNA) was amplified from viral RNA using the commercial available kit (Fermentas, USA) and based on the recommended instructions. Upon cDNA amplification, nested-PCR was performed for all the samples, to ensure the cDNA presence and their qualification. Then, optimized PCR was performed to amplify NS3 protease region. Since Max Taq enzyme (Vivantis Inc; Malaysia) and two types of buffer were used, PCR was performed in 2 stages using 2 different concentrations of buffers. First, amplification was performed using Max Taq enzyme in 1.6x concentration of buffer with the following steps: first round, 95°C for 5 minutes, then, 30 cycles of denaturation at 94°C for 50 seconds, annealing at 55°C for 45 seconds and extension at 72°C for 50 seconds; second round, PCR was performed in buffer A in 30 cycles by touch down program with annealing range of 53°C-59°C. Each annealing cycles was done in 45 seconds and finally, extension step was performed at 72°C for 40 seconds. In the end, the desired product sequences were extracted from agarose gel, in accordance with the manual of gel extraction kit (Invitek Inc, Germany).
3.4. Cloning and Sequencing
In this step, the extracted PCR product was cloned using TA/cloning system kit containing linear vector PTZ57R/T (Fermentas Inc., USA). Following the kit instructions, ligation was performed using T4 ligase enzyme at 37°C. Then, the reaction product was transformed into competent DH5α bacteria. The bacteria were grown on LB agar, containing ampicillin (50 µg/mL) along with X-Gal and IPTG. Recombinant true plasmids were initially screened by blue/white screening, and then confirmed by colony-PCR assay. For this purpose, colonies were cultured in LB broth media overnight, then, lysed at 95°C for 4 minutes, centrifuged at 12000 rpm and, finally, 2 µL of supernatant was introduced into PCR reaction tubes. Purification of true recombinant plasmids was done using Miniprep kit, based on the recommended instruction (Invitek Company, Germany). For each patient, nearly 6 colonies of true plasmids were sent for sequencing (Bioneer Inc., South Korea), which was performed bidirectional using F2G1b/F2G1a and Rev-2 (Common) primer set.
3.5. Sequence Analysis and Phylogeny
The NS3 sequence of different reference genotypes of HCV was retrieved from NCBI database, then, aligned using BioEdit and MEGA4 softwares. The employed reference sequences accession numbers for further analysis were as follows: genotype 1a, NC004102, D10749 and DQ068207; genotype 1b, AY460204, U01214, and AF176573; genotype 1C, AY651061 and D14853; genotype 2, AY746460 and AF169005; genotype 3a, D28917; genotype 4, Y11604; genotype 5, AF064490; and genotype 6, Y12083.
The sequencing results were retrieved through Chromas software, and then aligned by Clustal X software through comparing with reference sequences. Phylogenic tree was constructed by neighbor joining method using MEGA4 software. Finally, the created tree was evaluated by 1000 times bootstrapping to ensure its fidelity. During protease sequences, detailed survey substitutions of the amino acid residues of V36, T54, V55, Q80, S122, R155, A156, V158, D168, and I/V170 were considered.
4. Results
4.1. NS3 Protease Amplification and Cloning Results
To screen the presence of virus genome inside the samples, an in-house nested PCR was employed to amplify 5´NTR region. All the samples were found to be positive for HCV infection. A band of 247 bp was indicative of the presence of the specific amplicons in corresponding PCR reactions (Figure 1 A). All 10 patients’ samples were included in this step to amplify NS3 protease domain. Nested PCR was performed on the corresponding cDNA. Finally, a fragment with 744 bp was amplified in 7 out of 10 patients as depicted in Figure 1 B. The products of the second round were processed into gel extraction, purification, and cloning steps.
The Results of In-House Nested PCR and NS3 Amplification
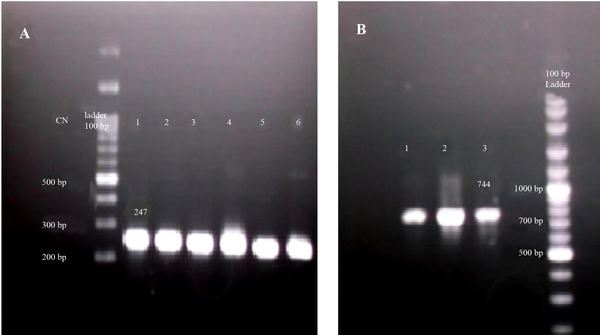
After ligation step, colony PCR assay was done to screen positive recombinant plasmids, containing NS3 protease domain. According to colony PCR results, about 90% of the white colonies were recombinant positive samples and nearly, all blue colonies were negative. Thus, nearly 6 white/positive colonies were submitted for sequencing with more than 90% success in sequencing step.
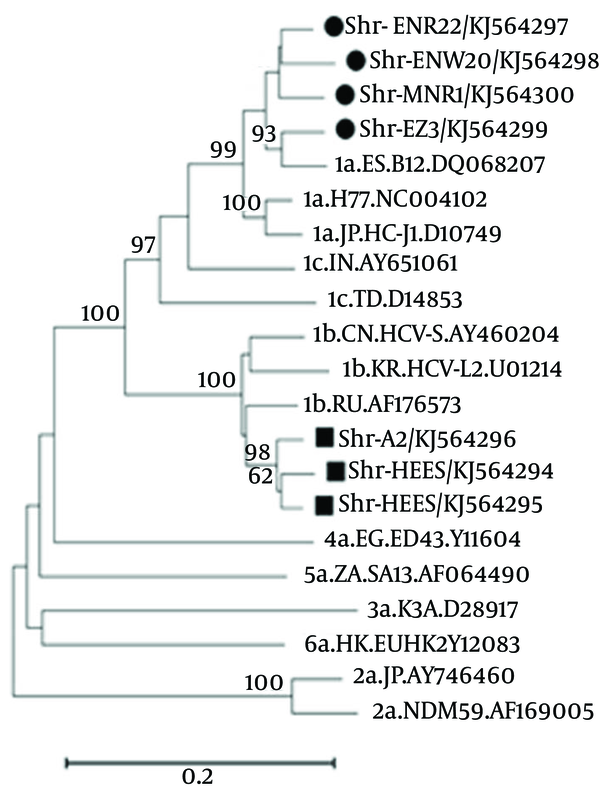
4.2. Sequencing and Phylogenic Analysis
All sequencing results for each patient were collected and consensus sequences were retrieved after their alignment. These consensus sequences were submitted to NCBI GeneBank with accession numbers of KJ564294 to KJ564300. The constructed phylogenic tree demonstrated that all our sample sequences are placed in the branch of genotype 1a or 1b, as shown in Figure 2. Of 7 analyzed cases, there were four KJ564297-300 and three KJ564294-96 samples with the genotypes 1a and 1b, respectively.
The Result of Phylogenic Tree
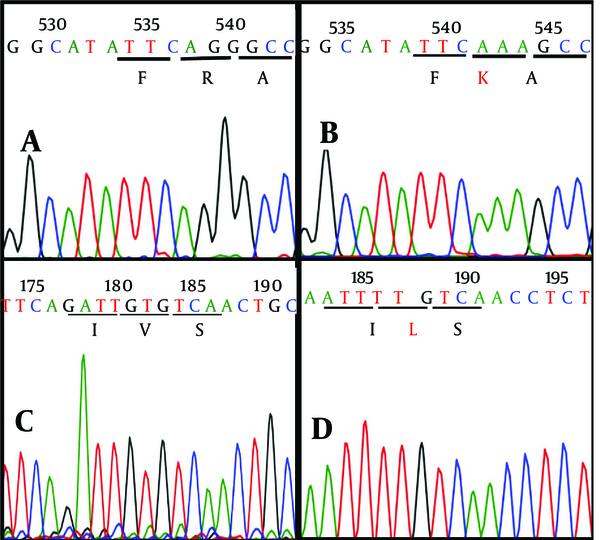
As the most important step, sequences were checked one by one to screen critical characterized point mutations and variations in nucleotide and amino acid sequences. Surprisingly, 2 critical mutations, including R155K (replacing of lysine by arginine in the position 155 aa) and V36L (valine to leucine substitution in the position 36 aa) were revealed in one (Shr-MNR1-KJ564300) out of 7 patients (Figures 3 and 4).
Detection of Critical Mutations in One Patient (Nominated as MNR1) During Screening by Comparison With the Reference Sequences

Furthermore, evaluation of the patient’s colonies demonstrated that R155K mutation was not dominant and detected only in two cloned plasmids while V36L mutation was diagnosed in all plasmid clones. The sequencing graphs were precisely supportive of this finding as depicted in Figure 4. Interestingly, crude sequencing result of this patient demonstrated solely V36L mutation, whereas R155K mutation remained undetectable. Therefore, it seems that cloning together with sequencing work properly and better than simple crude sequencing protocol.
Comparison of Sequencing Results From Wild and Mutant Viruses
The patient was a 50-year-old man who had experienced HCV relapse 1 year after receiving a course of standard therapy. It was also claimed that the source of infection came from his brother, as reported in his medical records. The brother had been infected with HCV genotype 1a but already obtained the sustained virology response following 1 year of therapy. In the mentioned sample, a new A39S variation was also recognized as a difference with all other HCV genotypes. Besides, S122G variation was detected in this case and sample KJ564299 too.
5. Discussion
Similar to other RNA viruses, extreme genetic diversity of HCV virus spontaneously creates new mutations that consequently impede novel drug functions (7, 10, 15). The discovery of a wide spectrum of protease inhibitors (PIs) is a major breakthrough in the treatment of HCV infection (16). Although the majority of studies showed the potential of PIs in resolving failures of standard treatment (17), emergence of natural drug resistance is predicted to be a challenging obstacle in this respect (7, 14). Recent experiments have revealed that natural genetic polymorphisms and resistant mutants to PIs inside the NS3 protease domain exist even in the patients who do not receive the drugs (7, 13, 14). Unfortunately, some of these mutations induce resistance to all established PIs, a tragedy for virus harboring patients. Among these crucial mutation, R155K have a special place in inducing telaprevir, boceprevir, and other protease inhibitors resistance, drugs approved by the FDA and available in the market now.
Dozens of experiments reported the rate of natural mutation for NS3 protease in different areas (14, 18, 19). In a pioneer study, the heterogeneity of the NS3 protease region in 17 patients with genotype-1, was evaluated by sequencing and a significant level of heterogeneity was confirmed along the gene (12). In one of the most comprehensive studies, a significant proportion (close to 8%) of the drug resistant mutations for PIs was reported among a big population (14). After the initial assessment, the screening for well-characterized PIs mutations was recommended before starting the treatment. This strategy consequently reduces the cost and prevents treatment failure (4, 14). High prevalence of resistance mutation to PIs in different genotypes were also reported elsewhere (20). However, a report from France detected no HCV case harboring critical R155K/T/M, A156S/V/T, and V170A when screening accomplished in 108 genotype 1 patients by direct sequencing (21). Recently, Trevino et al. conducted a study on drug resistance in HCV-HIV co-infected patients by direct sequencing, but they did not recognize R155 mutation (22). Detection of R155K variant is a rare event usually happens during large population screening (23).
After the availability of PIs drugs in the market, natural mutation still remains a big concern, especially in developing countries where such drugs are not accessible enough and are costly. The present preliminary study sought to evaluate natural PIs resistant mutations in a randomly small selected patient population, referred to a central liver center in south of Iran. Also, instead of conventional sequencing, clonal-sequencing was performed to improve the mutation detection limit. It seems that employment of direct sequencing method can reduce the sensitivity of detection and consequent underestimation (21). The obtained sequences were analyzed with bioinformatics software and the phylogenetic tree showed the presence of all selected sequences in genotype 1a or 1b branch.
Interestingly, crucial mutation of R155K and V36L were detected in one of the studied patients. Furthermore, the R155K mutation was found only in 2 clones, whereas V36L mutation in all clones, as the dominant mutation. Detection of critical R155 mutation in such a small population is so alarming that it requires further investigations with a larger population. R155K and V36L mutations are very critical and can cause drug resistance to a wide range of protease inhibitors. In our mutated sample, a new A39S variation was also detected that was different from all other HCV genotypes. The exact role of this variation is not clear in the literature and it may just act as a polymorphism or compensatory mutation (24). Besides, S122G variation that may responsible for resistance to one protease inhibitor detected in two samples (mutated case and another sample). However, its impact on the course of treatment underestimated in recent publications (23, 24).
Detailed investigations of protease region for natural mutations before starting the widespread treatment with PIs can contribute to greater treatment success rate and avoid waste of money. Such investigations also help selection of suitable PIs for the respective patients. Epidemiologically, the HCV mutant patient was claimed to develop infection through his brother, as reported in his medical records. Even though both genotypes were 1a, however, it is unknown if HCV R155K mutated virus was transmitted by family source or was gradually developed in the patient. Further molecular epidemiology was impossible because of sustained virology response of brother and undetectable virus level.
In conclusion, in this study on 7 patients, the first HCV infected case with R155K natural resistance to PIs drugs in south of Iran was detected. By estimation and considering the random process of selection, it seems that the rate of PIs natural mutants is considerable in our HCV infected patients.
Acknowledgements
References
-
1.
Alavian SM, Adibi P, Zali MR. Hepatitis C virus in Iran: Epidemiology of an emerging infection. Arch Iran Med. 2005;8(2):84-90.
-
2.
Vermehren J, Sarrazin C. New HCV therapies on the horizon. Clin Microbiol Infect. 2011;17(2):122-34. [PubMed ID: 21087349]. https://doi.org/10.1111/j.1469-0691.2010.03430.x.
-
3.
Courcambeck J, Bouzidi M, Perbost R, Jouirou B, Amrani N, Cacoub P, et al. Resistance of hepatitis C virus to NS3-4A protease inhibitors: mechanisms of drug resistance induced by R155Q, A156T, D168A and D168V mutations. Antivir Ther. 2006;11(7):847-55. [PubMed ID: 17302247].
-
4.
Kim AY, Timm J, Nolan BE, Reyor LL, Kane K, Berical AC, et al. Temporal dynamics of a predominant protease inhibitor-resistance mutation in a treatment-naive, hepatitis C virus-infected individual. J Infect Dis. 2009;199(5):737-41. [PubMed ID: 19199553]. https://doi.org/10.1086/596657.
-
5.
Parfieniuk A, Jaroszewicz J, Flisiak R. Specifically targeted antiviral therapy for hepatitis C virus. World J Gastroenterol. 2007;13(43):5673-81. [PubMed ID: 17963291].
-
6.
Kieffer TL, Kwong AD, Picchio GR. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J Antimicrob Chemother. 2010;65(2):202-12. [PubMed ID: 19903720]. https://doi.org/10.1093/jac/dkp388.
-
7.
Romano KP, Ali A, Royer WE, Schiffer CA. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc Natl Acad Sci U S A. 2010;107(49):20986-91. [PubMed ID: 21084633]. https://doi.org/10.1073/pnas.1006370107.
-
8.
Fields B, Knipe D, Howley PM. Fields virology. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2013.
-
9.
Mayhoub AS. Hepatitis C RNA-dependent RNA polymerase inhibitors: a review of structure-activity and resistance relationships; different scaffolds and mutations. Bioorg Med Chem. 2012;20(10):3150-61. [PubMed ID: 22516671]. https://doi.org/10.1016/j.bmc.2012.03.049.
-
10.
Le Guillou-Guillemette H, Vallet S, Gaudy-Graffin C, Payan C, Pivert A, Goudeau A, et al. Genetic diversity of the hepatitis C virus: impact and issues in the antiviral therapy. World J Gastroenterol. 2007;13(17):2416-26. [PubMed ID: 17552024].
-
11.
Neau D, Jouvencel AC, Legrand E, Trimoulet P, Galperine T, Chitty I, et al. Hepatitis C virus genetic variability in 52 human immunodeficiency virus-coinfected patients. J Med Virol. 2003;71(1):41-8. [PubMed ID: 12858407]. https://doi.org/10.1002/jmv.10451.
-
12.
Vallet S, Gouriou S, Nousbaum JB, Legrand-Quillien MC, Goudeau A, Picard B. Genetic heterogeneity of the NS3 protease gene in hepatitis C virus genotype 1 from untreated infected patients. J Med Virol. 2005;75(4):528-37. [PubMed ID: 15714495]. https://doi.org/10.1002/jmv.20302.
-
13.
Lopez-Labrador FX, Moya A, Gonzalez-Candelas F. Mapping natural polymorphisms of hepatitis C virus NS3/4A protease and antiviral resistance to inhibitors in worldwide isolates. Antivir Ther. 2008;13(4):481-94. [PubMed ID: 18672527].
-
14.
Kuntzen T, Timm J, Berical A, Lennon N, Berlin AM, Young SK, et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology. 2008;48(6):1769-78. [PubMed ID: 19026009]. https://doi.org/10.1002/hep.22549.
-
15.
Paolucci S, Fiorina L, Mariani B, Gulminetti R, Novati S, Barbarini G, et al. Naturally occurring resistance mutations to inhibitors of HCV NS5A region and NS5B polymerase in DAA treatment-naive patients. Virol J. 2013;10:355. [PubMed ID: 24341898]. https://doi.org/10.1186/1743-422X-10-355.
-
16.
Zeminian LB, Padovani JL, Corvino SM, Silva GF, Pardini MI, Grotto RM. Variability and resistance mutations in the hepatitis C virus NS3 protease in patients not treated with protease inhibitors. Mem Inst Oswaldo Cruz. 2013;108(1):13-7. [PubMed ID: 23440108].
-
17.
Winters MA, Chary A, Eison R, Asmuth D, Holodniy M. Impact of highly active antiretroviral therapy on hepatitis C virus protease quasispecies diversity in HIV co-infected patients. J Med Virol. 2010;82(5):791-8. [PubMed ID: 20336744]. https://doi.org/10.1002/jmv.21679.
-
18.
Vicenti I, Rosi A, Saladini F, Meini G, Pippi F, Rossetti B, et al. Naturally occurring hepatitis C virus (HCV) NS3/4A protease inhibitor resistance-related mutations in HCV genotype 1-infected subjects in Italy. J Antimicrob Chemother. 2012;67(4):984-7. [PubMed ID: 22258932]. https://doi.org/10.1093/jac/dkr581.
-
19.
Colson P, Brouk N, Lembo F, Castellani P, Tamalet C, Gerolami R. Natural presence of substitution R155K within hepatitis C virus NS3 protease from a treatment-naive chronically infected patient. Hepatology. 2008;47(2):766-7. [PubMed ID: 18220300]. https://doi.org/10.1002/hep.22122.
-
20.
Peres-da-Silva A, de Almeida AJ, Lampe E. Mutations in hepatitis C virus NS3 protease domain associated with resistance to specific protease inhibitors in antiviral therapy naive patients. Arch Virol. 2010;155(5):807-11. [PubMed ID: 20405151]. https://doi.org/10.1007/s00705-010-0642-z.
-
21.
Vallet S, Viron F, Henquell C, Le Guillou-Guillemette H, Lagathu G, Abravanel F, et al. NS3 protease polymorphism and natural resistance to protease inhibitors in French patients infected with HCV genotypes 1-5. Antivir Ther. 2011;16(7):1093-102. [PubMed ID: 22024525]. https://doi.org/10.3851/IMP1900.
-
22.
Trevino A, de Mendoza C, Parra P, Rodriguez C, Madejon A, Plaza Z, et al. Natural polymorphisms associated with resistance to new antivirals against HCV in newly diagnosed HIV-HCV-coinfected patients. Antivir Ther. 2011;16(3):413-6. [PubMed ID: 21555824]. https://doi.org/10.3851/IMP1760.
-
23.
Alexopoulou A, Karayiannis P. Interferon-based combination treatment for chronic hepatitis C in the era of direct acting antivirals. Ann Gastroenterol. 2015;28(1):55-65. [PubMed ID: 25608803].
-
24.
Paolucci S, Fiorina L, Piralla A, Gulminetti R, Novati S, Barbarini G, et al. Naturally occurring mutations to HCV protease inhibitors in treatment-naive patients. Virol J. 2012;9:245. [PubMed ID: 23095680]. https://doi.org/10.1186/1743-422X-9-245.