Abstract
Background:
Candida species are usually opportunistic organisms that cause acute to chronic infections when conditions in the host are favorable. Accurate identification of Candida species is an essential pre-requisite for improved therapeutic strategy. Identification of Candida species by conventional methods is time-consuming with low sensitivity, yet molecular approaches have provided an alternative way for early diagnosis of invasive candidiasis. Denaturing gradient gel electrophoresis (DGGE) and temporal temperature gradient gel electrophoresis (TTGE) are polymerase chain reaction (PCR)-based approaches that are used for studying the community structure of microorganisms. By using these methods, simultaneous identification of multiple yeast species will be possible and reliable results will be obtained quickly.Objectives:
In this study, DGGE and TTGE methods were set up and evaluated for the detection of different Candida species, and their results were compared.Materials and Methods:
Five different Candida species were cultured on potato dextrose agar medium for 24 hours. Next, total DNA was extracted by the phenol-chloroform method. Two sets of primers, ITS3-GC/ITS4 and NL1-GC/LS2 were applied to amplify the desired regions. The amplified fragments were then used to analyze DGGE and TTGE profiles.Results:
The results showed that NL1-GC/LS2 primer set could yield species-specific amplicons, which were well distinguished and allowed better species discrimination than that generated by the ITS3-GC/ITS4 primer set, in both DGGE and TTGE profiles. All five Candida species were discriminated by DGGE and TTGE using the NL1-GC/LS2 primer set.Conclusions:
Comparison of DGGE and TTGE profiles obtained from NL1-GC/LS2 amplicons exhibited the same patterns. Although both DGGE and TTGE techniques are capable of detecting Candida species, TTGE is recommended because of easier performance and lower costs.Keywords
Molecular Assay Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis Polymerase Chain Reaction-Temporal Temperature Gradient Gel Electrophoresis Candida
1. Background
Candida species are the most common cause of fungal infections, leading to a range of life threatening and non-life-threatening diseases. Because of widespread use of broad-spectrum antibiotics and the growing numbers of HIV-infected and immune-compromised patients, the incidence of Candida infections has grown in the recent years (1). Vaginal candidiasis causes 20 - 25% of infectious vaginitis cases (2). It is estimated that 75% of all females experience an episode of vaginal candidiasis in their lifetime (3). Candida albicans, C. glabrata, C. parapsilosis, and C. tropicalis account for 80 - 90% of fungal isolates that cause infections worldwide (4). Accurate identification of Candida species is important for the treatment of infected patients, because significant attributes such as drug resistance and virulence differ among species (5).
Commonly, Candida in vaginal samples is identified by microscopic examination of a wet mount with potassium hydroxide (KOH, amine test). Using this technique budding yeast cells in only 50 - 70% of females with Candida vaginitis are detected and non-albicans species may not be identified (6). Alternatively, C. albicans and C. tropicalis can be distinguished by growth on chromogenic agar medium and other Candida species can be recognized by enzymatic tests. For each of these tests, isolated organisms have to be grown on solid medium for 24 to 48 hours before the test. In addition, the ‘gold standard’ for definitive biochemical identification requires analysis of assimilation and fermentation, taking up to 30 days to complete (7).
In the recent years, numerous DNA-based techniques have been developed to improve the identification of Candida species and provide more accurate results. These molecular techniques include the polymerase chain reaction (PCR), real-time PCR, restriction fragment length polymorphism, electrophoretic karyotyping, fluorescence in situ hybridization, randomly amplified polymorphic DNA analysis, multi-locus sequence typing and pyrosequencing. Each technique has its own advantages and limitations, and many of them require a culturing step to isolate the target species (8). Therefore, culture bias and the loss of minor species can occur. Real-time PCR method has been developed to detect medically important Candida species in clinical samples (9). This novel method is sensitive, specific and rapid for Candida detection and estimation, yet it also has its own drawbacks. The main limitation is its specificity for particular species or groups of species in a situation where multiple or unpredicted species may occur.
The use of PCR together with denaturing gradient gel electrophoresis (DGGE) or the related method temporal temperature gradient gel electrophoresis (TTGE) enables detection of the presence of such species and frequently their presumptive identification, even if they are present as minor populations. In both TTGE and DGGE, DNA fragments of the same length yet with different sequences are separated, based on decreased electrophoretic mobility of partially melted double-strand DNA molecules. Separation is performed with polyacrylamide gels containing a linear gradient of chemical denaturant gradient in DGGE or a linear temperature gradient in TTGE (10).
2. Objectives
In this study, the ability of DGGE and TTGE techniques to distinguish different Candida species was assayed and the results of profiles were compared.
3. Materials and Methods
3.1. Samples and DNA Extraction
Five different standard Candida species including C. albicans (CCUG 32723), C. glabrata (CCUG 35267), C. tropicalis (CCUG 34274), C. orthopsilosis (CCUG 20503), C. parapsilosis (ATTC 22019), and some clinical strains isolated from vaginal tracts were analyzed in this study. These strains were cultured on Potato Dextrose Agar, PDA (Merck, Germany) for 24 hours at 36°C and their genomic DNA was extracted using the phenol-chloroform method, according to isolation of nucleic acid protocols of yeast (11).
3.2. Polymerase Chain Reaction Conditions
All PCR reactions were performed in a PeqSTAR thermocycler (PEQLAB, Germany). Two primer sets were used. The first primer set was ITS3 and ITS4 (12), and 40 bp GC-clamp was attached to the 5’ end of the forward primer (13). This primer set amplifies the ITS2 region, producing amplicons with ~ 300 - 400 bp length. The second primer set was NL1/LS2 and the 30 bp GC clamp was attached to the 5’ end of NL1. This primer set amplifies the D1 region of the 26 - 28S rRNA gene, yielding amplicons of ~ 250 bp length (14). For the first PCR reactions, 5 μL of PCR buffer (CinnaGen, Iran), 1.5 mM of MgCl2 (CinnaGen, Iran), 0.2 mM of dNTPs (CinnaGen, Iran), 0.16 mM of each primer (Faza Biotech, Iran), 1.25 U of DNA Taq polymerase (CinnaGen, Iran) and about 20 ng of DNA were used in a final volume of 50 µL. The amplification program of DNA was started with denaturation for five minutes at 95°C, followed by 35 cycles of denaturation for 30 seconds at 95°C, primer annealing for 45 seconds at 58°C and extension for one minute at 72°C. Final extension was at 72°C for five minutes. For the second PCR, the 50 µL reaction mixture consisted of 5 µL of PCR buffer (CinnaGen, Iran) supplemented with 4 mM MgCl2 (CinnaGen, Iran), 0.2 mM dNTPs (CinnaGen, Iran), 0.1 mM of each primer (Faza Biotech, Iran), 2.5 U Taq DNA polymerase (CinnaGen, Iran) and about 20 ng of DNA. The reactions were performed for 30 cycles. Following an initial four-minute denaturation at 95°C, the PCR cycle consisted of 95°C for 30 seconds, 53°C for 45 seconds and 72°C for 60 seconds, with a final extension at 72°C for seven minutes. Polymerase Chain Reaction products (5 µL) were analyzed on 1% (w/v) agarose gel (Merck, Germany) by electrophoresis (Bio-Rad, USA).
3.3. Denaturing Gradient Gel Electrophoresis and Temporal Temperature Gradient Gel Electrophoresis Conditions
The DCode universal mutation detection system (Bio-Rad, Hercules, CA, USA) was used for DGGE and TTGE analysis. Denaturing Gradient Gel Electrophoresis was performed using a 1.0 mm polyacrylamide gel [ratio of 8% (w/v) acrylamide (Merck, Germany) to bis-acrylamide (Merck, Germany), 37.5: 1]. For the ITS3-GC/ITS4 primer PCR products, 30 - 60% denaturing gradient [100% denaturant was 7 M urea (Merck, Germany), and 40% (v/v) formamide (Merck, Germany)], which increased in the direction of electrophoresis, was applied. The gel was prepared and run with 1X Tris-acetate-EDTA (TAE) buffer at a constant voltage of 55 V at 56°C for 16 hours. For primer set NL1-GC/LS2 amplicons, DGGE was performed with a 30 - 45% denaturing gradient and run with 1X TAE at a constant voltage of 130 V for 4.5 hours at 60°C.
In TTGE, the polyacrylamide gel composed of 8% (v/v) acrylamide-bis-acrylamide mixture. Temporal temperature gradient gel electrophoresis was performed at a constant voltage of 65 V and 6 M urea (Merck, Germany) for 14 hours and 17 minutes with a temperature gradient of 56 to 66°C for the ITS2 region and at a constant voltage of 65 V and 7 M urea for 10 hours and 42 minutes with a temperature gradient of 51/5 to 62/2°C for the D1 region. The PCR products (20 μL) were mixed with 20 μL 2x gel loading dye. After electrophoresis, the gels were stained with ethidium bromide (Merck, Germany) for 30 minutes at room temperature and visualized by a UV transillumination (Syngen inGenius LHR, UK).
4. Results
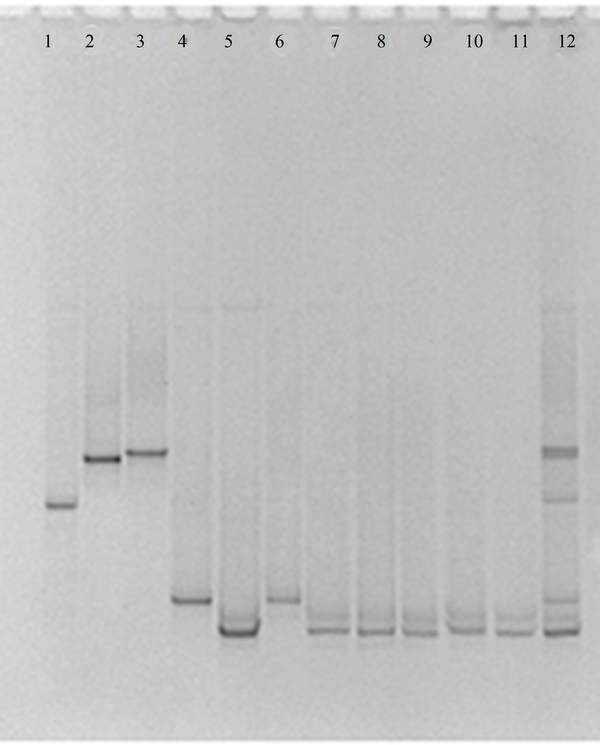
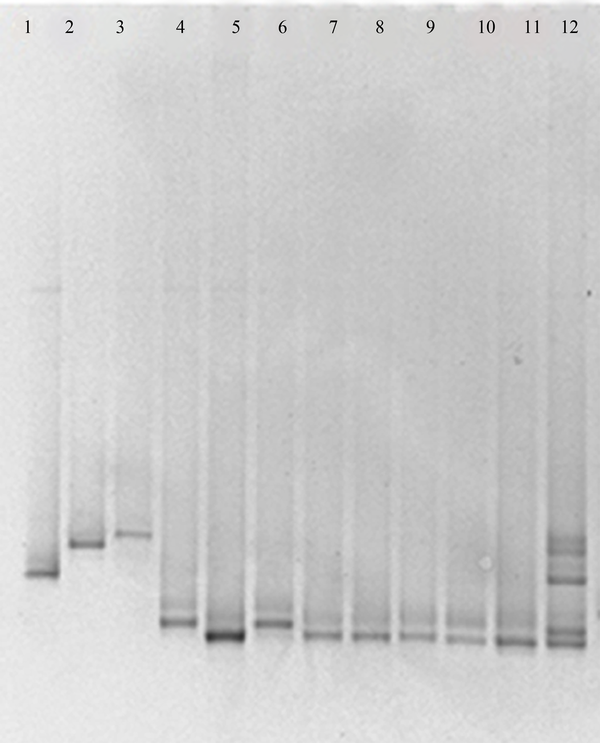
Using the ITS3-GC/ITS4 primer pair for the ITS2 region, unspecific PCR products were produced, which were not eliminated by changing PCR conditions, such as number of cycles, annealing temperature, primer concentration or amount of template. By using these PCR products in DGGE and TTGE, appropriate results were not found. Multiple bands were visualized in the fingerprints derived from a single Candida species and some of species were not discriminated from each other (data not shown). The NL1-GC/LS2 primer set produced specific PCR products and by applying these products in DGGE and TTGE for each Candida species, reproducible clear single bands at a specific position of the gel were achieved (Figures 1 and 2). By comparing clinical samples with standard species, isolated yeasts were identified as C. albicans and C. glabrata.
Denaturing Gradient Gel Electrophoresis Profiles Obtained From Candida Species With the NL1-Gc/Ls2 Primer Set
Temporal Temperature Gradient Gel Electrophoresis Profiles Obtained From Candida Species With the NL1-Gc/Ls2 Primer Set
5. Discussion
The incidence of opportunistic Candida infections has considerably increased in the recent years. Despite progress in the development of new molecular approaches for the diagnosis of Candida infections, developing a simple, rapid, and economic effective test for diagnostic purposes remains elusive. In the present study, DGGE and TTGE techniques were evaluated to detect different Candida species by using two primer sets, ITS3-GC/ITS4 and NL1-GC/LS2. These methods are rapid, inexpensive and reliable and allow simultaneous analysis of multiple samples on the same gel (15). Furthermore, DGGE and TTGE can be used in clinical laboratories to identify pathogenic Candida species. Weerasekera et al. used the DGGE method for the identification of various Candida species (16) yet the TTGE technique has not been used for detection of Candida species so far.
Our results showed that the ITS3-GC/ITS4 primer pair was not able to produce appropriate PCR products and yielded multiple banding patterns for some of the species being analyzed by DGGE and TTGE. These findings are in accordance with other studies, which were carried out using ITS3-GC/ITS4 primers (17). As the internal transcribed spacer (ITS) region is highly variable, production of multiple bands in DGGE and TTGE may be due to the sequence variation between different ITS2 copies in the rRNA operon. Therefore, using the ITS3-GC/ ITS4 primer pair in DGGE and TTGE may lead to overestimation of microorganism populations being analyzed (18).
Separation of DNA fragments in DGGE and TTGE is based on decreased electrophoretic mobility of partially melted double-stranded DNA on polyacrylamide gels containing a linear gradient of DNA denaturant in DGGE or a linear temperature gradient in TTGE. Moreover, DGGE is complicated by the difficulties of choosing the exact running time and gel contents to achieve optimal separation. By running a DGGE gel for too long, an achieved separation decreases, and may even be lost. Furthermore, gel-casting conditions are not exactly reproducible in DGGE and it is not possible to compare two different gels. In contrast, in TTGE, the denaturant concentration and running time can be determined from theoretical melting curves. Since there is no denaturant gradient, TTGE gel has higher reproducibility in casting, and easier preparation (19). As it has been shown in the current study by increasing voltage in DGGE, the electrophoresis time will be reduced. On the other hand, in TTGE, the electrophoresis time depends on ramp rate (temperature increase °C/hour). By increasing ramp rate to more than 1°C/hour, band resolution decreases. Therefore, in the extensive temperature ranges, the required time for TTGE is more than DDGE.
In the current study comparison of DGGE and TTGE profiles showed that both of these techniques are capable to differentiate various Candida species, however, in the study that was carried out by Farnleitner et al. results showed that TTGE is not able to separate DNA amplicons from each other while DGGE was able to do so (20). Our study exhibited that bands migration for different Candida species has the same patterns in both DGGE and TTGE profiles, yet resolution of bands in DGGE was better in comparison with TTGE. As both DGGE and TTGE methods are based on the same principles, it is expected for their results to follow the same patterns. However, Marie et al. reported that DGGE and TTGE patterns appear not to be directly comparable in the study of picoplankton communities of the Mediterranean Sea (21).
Our data suggested that by using the NL1-GC-LS2 primer pair, both of DGGE and TTGE techniques are capable to detect different Candida species, and although TTGE takes a long time, this method is more appropriate than DGGE for Candida species due to lower cost, simplicity and reproducibility.
Acknowledgements
References
-
1.
Jarvis WR. Epidemiology of nosocomial fungal infections, with emphasis on Candida species. Clin Infect Dis. 1995;20(6):1526-30. [PubMed ID: 7548503].
-
2.
Trama JP, Mordechai E, Adelson ME. Detection and identification of Candida species associated with Candida vaginitis by real-time PCR and pyrosequencing. Mol Cell Probes. 2005;19(2):145-52. [PubMed ID: 15680216]. https://doi.org/10.1016/j.mcp.2004.10.004.
-
3.
Sobel JD. Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann N Y Acad Sci. 1988;544:547-57. [PubMed ID: 3063184].
-
4.
Pfaller MA. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis. 1996;22 Suppl 2:S89-94. [PubMed ID: 8722834].
-
5.
Lopez-Martinez R. Candidosis, a new challenge. Clin Dermatol. 2010;28(2):178-84. [PubMed ID: 20347660]. https://doi.org/10.1016/j.clindermatol.2009.12.014.
-
6.
Elliott KA. Managing patients with vulvovaginal candidiasis. Nurse Pract. 1998;23(3):44-6. [PubMed ID: 9560829].
-
7.
Warren NG, Hazen KC. Candida, Cryptococcus, and other yeasts of medical importance. In: Murray PR, Barton EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of clinical microbiology. Washington, DC: American Society for Microbiology; 1995. 723–37 p.
-
8.
Trtkova J, Raclavsky V. Molecular-genetic approaches to identification and typing of pathogenic Candida yeasts. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150(1):51-61. [PubMed ID: 16936901].
-
9.
Fricke S, Fricke C, Schimmelpfennig C, Oelkrug C, Schonfelder U, Blatz R, et al. A real-time PCR assay for the differentiation of Candida species. J Appl Microbiol. 2010;109(4):1150-8. [PubMed ID: 20456528]. https://doi.org/10.1111/j.1365-2672.2010.04736.x.
-
10.
Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek. 1998;73(1):127-41. [PubMed ID: 9602286].
-
11.
Xiano W. Yeast Protocols. In: Hanna M, Xiano W, editors. Isolation of Nucleic Acids. Berlin: springer; 2006. p. 2-4.
-
12.
White TJ, Bruns T, Lee S, Taylor J. Amplification and Direct Sequencing of Fungal Ribosomal Rna Genes for Phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. 315–22 p. https://doi.org/10.1016/b978-0-12-372180-8.50042-1.
-
13.
Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695-700. [PubMed ID: 7683183].
-
14.
Cocolin L, Bisson LF, Mills DA. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett. 2000;189(1):81-7. [PubMed ID: 10913870].
-
15.
Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol. 1999;2(3):317-22. [PubMed ID: 10383868]. https://doi.org/10.1016/S1369-5274(99)80055-1.
-
16.
Weerasekera MM, Sissons CH, Wong L, Anderson S, Holmes AR, Cannon RD. Use of denaturing gradient gel electrophoresis for the identification of mixed oral yeasts in human saliva. J Med Microbiol. 2013;62(Pt 2):319-30. [PubMed ID: 23065546]. https://doi.org/10.1099/jmm.0.050237-0.
-
17.
Michaelsen A, Pinzari F, Ripka K, Lubitz W, Pinar G. Application of molecular techniques for identification of fungal communities colonising paper material. Int Biodeter Biodegr. 2006;58(3-4):133-41. https://doi.org/10.1016/j.ibiod.2006.06.019.
-
18.
Kowalchuk G, Smit E. Fungal community analysis using PCR-denaturing gradient gel electrophoresis (DGGE). In: Kowalchuk GA, Bruijn FJ, Head IM, Akkermans ADL, vanElsas JD, editors. Molecular microbial ecology manual. Dordrech: Kluwer Academic Publishers; 2004. 771–88 p.
-
19.
Borresen-Dale AL, Lystad S, Langeroed A. Temporal temperature gradient electrophoresis on the DCode system. Biorad Bull. 1997;8:2133.
-
20.
Farnleitner AH, Kreuzinger N, Kavka GG, Grillenberger S, Rath J, Mach RL. Comparative analysis of denaturing gradient gel electrophoresis and temporal temperature gradient gel electrophoresis in separating Escherichia coli uidA amplicons differing in single base substitutions. Lett Appl Microbiol. 2000;30(6):427-31. [PubMed ID: 10849270].
-
21.
Marie D, Zhu F, Balague V, Ras J, Vaulot D. Eukaryotic picoplankton communities of the Mediterranean Sea in summer assessed by molecular approaches (DGGE, TTGE, QPCR). FEMS Microbiol Ecol. 2006;55(3):403-15. [PubMed ID: 16466379]. https://doi.org/10.1111/j.1574-6941.2005.00058.x.