Abstract
Background:
Pseudomonas aeruginosa is a well-known opportunistic pathogen, which affects hospitalized patients in different wards due to its natural resistance to drugs.Objectives:
The purpose of the current study was to determine the antibiotic susceptibility profiles and genetic relatedness in P. aeruginosa isolated from patients admitted to a referral hospital in Isfahan, Iran.Materials and Methods:
Out of 150 analyzed samples, 54 P. aeruginosa isolates were recovered and were subjected to antibiotic resistance patterns and genetic diversity determination by Kirby-Bauer’s disk diffusion method and RAPD-PCR, respectively.Results:
The highest percentage of resistance was observed against ceftazidime and imipenem with 30 (55.6%) isolates; meanwhile all isolates were sensitive to polymyxin B. Twenty-eight (51.8%) isolates revealed resistance to all applied antibiotics. RAPD-PCR (Random Amplified Polymorphic DNA- Polymerase Chain Reaction) results showed 54 unique genotypes, which were divided into 39 clusters.Conclusions:
Although different source of P. aeruginosa may involve in patient colonization, genetically related strains were isolated from different wards and or the same ward of the hospital. Our results pointed to the restriction of currently used antibiotics in studied hospital. We hope that our results cast light on the control and transmission of the infection in the investigated hospital.Keywords
1. Background
Pseudomonas aeruginosa is an important Gram-negative opportunistic pathogen that has the capability to create variable clinical infections, including, but not limited to, wound infections, urinary tract infections (UTIs), and blood stream infections (BSI), particularly in hospitalized and immunocompromised patients (1, 2). Pseudomonas aeruginosa was the second most common cause of hospital-acquired pneumonia, as well as the third and fifth common cause of hospital-acquired UTIs in USA and Europe, respectively (2-4). Evidently, P. aeruginosa due to low permeability of its outer membrane in combination with efflux pump overexpression is intrinsically resistant to a variety of antibiotics (5). Expression of different classes of extended-spectrum β-lactamas enzymes along with aminoglycoside-modifying enzymes (aminoglycoside phosphoryltransferases, aminoglycoside acetyltransferases, and aminoglycoside nucleotidyltransferases), and mutation are other well-characterized mechanisms for antibiotic resistance in P. aeruginosa (6, 7). Data extracted from several independent studies in Iran showed that P. aeruginosa was responsible for 11%-32.4% of UTIs, 17.2%-32.4% of pneumonia, 36.7% of BSI, and 47% of wound infections (8-12). Moreover, based on previous studies performed in Iran, treatment of P. aeruginosa infections is a major concern for health-care setting due to its high resistant rate to different antibiotics (12, 13).
Since determining bacterial isolates relatedness is essential for understanding the transmission routes, different typing methods that can be divided into two major categories, phenotypic and genotypic methods, have been established (14). Phenotypic methods such as serotyping, pyocin typing, and antimicrobial susceptibility typing depend on the environmental factors, therefore, have low discriminatory power (14, 15). Genotypic methods have been performed by different typing techniques such as Restriction Fragment Length Polymorphism (RFLP), Pulsed Field Gel Electrophoresis (PFGE), Multilocus Sequence Typing (MLST), Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR), Variable Number Tandem Repeat (VNTR), DNA Hybridization and Random Amplified Polymorphic DNA (RAPD) (16-21). Although MLST and PFGE have more discriminatory power, they are expensive, especially in developing countries (22). RAPD-PCR has the advantage of being fast, reproducible, simple, and low-cost, thus, it is an appropriate approach for primary screening of epidemic strains in large number of isolates (23, 24).
2. Objectives
To our knowledge, limited data are available about genetic diversity of P. aeruginosa recovered from different clinical specimens in Isfahan; the purpose of the current study was to determine the antibiotic susceptibility profiles and genetic relatedness in P. aeruginosa isolated from patients admitted to a referral hospital in Isfahan, Iran.
3. Materials and Methods
3.1. Pseudomonas aeruginosa Isolation and Identification
This study was conducted at a teaching referral hospital of Isfahan, Iran, from February 2013 to November 2013. During the mentioned period, a total of 150 non-duplicate samples (urine, sputum, wound, blood, and eye discharge) taken from hospitalized-patients admitted to different wards of hospital were analyzed. Primary identification of P. aeruginosa was done based on the standard conventional biochemical tests, including Gram staining, catalase, oxidase, oxidative-fermentative (OF) tests, pigment production and growth at 42°C. Then, primary identification was further confirmed with species-specific PCR using ITS (16s-23s rRNA internal transcribed spacer) –primer (25, 26). The study was approved by the Ethics Committee of Isfahan University of Medical Sciences (No. 392063).
3.2. Antibiotic Susceptibility Test
Resistance to antibiotics was evaluated by Kirby-Bauer’s disk diffusion method according to CLSI (Clinical Laboratory Standard Institute) recommendation (27). The following disks (MAST, UK) were applied: ceftazidime (CAZ, 30 µg), imipenem (IMP, 10 µg), meropenem (MEM, 10 µg), ciprofloxacin (CIP, 5 µg), aztreonam (ATM, 30 µg), polymyxin B (PB, 300 units), and amikacin (AMK, 30 µg). P. aeruginosa standard strain (ATCC 27853) was used as the quality control.
3.3. Random Amplified Polymorphic DNA-Genotyping
Optimized RAPD reactions mixtures comprised 2.5 µL 10x PCR buffer, 2.5 mM MgCl2, 300 µM of dNTPs, 1.7 U Taq DNA polymerase ( CinnaGen, Iran), 3 µL genomic DNA (40 ng), and 10 pM of 272- AGCGGGCCAA primer (21) (Metabion, Germany) in 25 µL final volume. DNA amplification was carried out using Biometra thermocycler (Germany) and following a two-step program, 1) denaturation 5 min at 95°C, annealing 5 min at 36°C, elongation 5 min at 72°C, for 4 cycles, and 2) 31 cycles consisted of 94°C for 1 min, 45°C for 1 min, 72°C for 2 min, followed by a final extension at 72°C for 10 min (21). Electrophoresis was carried out using 2% agarose gel (w/v) and 0.5x TBE (Tris-Boric acid-EDTA, pH = 7.5-8) buffer at 7 v/cm for 3h. We also used 100 bp DNA ladder (CinnaGen, Iran) as the standard molecular size (100 bp-3000 bp). Separated bands stained with ethidium bromide (0.5 µg/mL) and visualized picture was captured on Gel-documentation system (Uvitec, UK). RAPD-fingerprints were recorded as present (1) or absent (0) for each band. By means of FreeTree and TreeView softwares, Dice similarity coefficient and Unweighted Average Pair Group method (UPGMA) were used for similarity matrix calculation and cluster analysis, respectively (28, 29). Only major reproducible bands regardless of their intensity were considered for similarity matrix calculation (30). We used cut-off value of ≥ 80% for determination of potential clonal relatedness (31-33).
4. Results
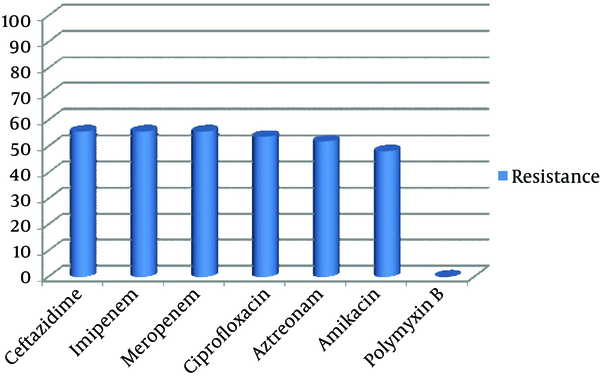
Out of 150 samples, 54 (36%) were positive for P. aeruginosa culture. Only one isolate per patient was recruited for study. Distribution of the 54 isolates were as follow, urine 39 (72.2 %), sputum 7 (13%), wound 5 (9.3%), blood 2 (3.7%), and eye discharge 1 (1.9%). The most frequent involved wards were ICU 28 (51.9%), followed by Graft 12 (22.2%), internal 5 (9.3%) and Surgery 9 (16.7%). Thirty-six isolates (66.7%) belonged to male patients and 18 (33.3%) belonged to female patients. The highest percentage (55.6%) of resistance was observed against ceftazidime and imipenem with 30 of the isolates being resistant; all isolates were sensitive to polymyxin B (Figure 1). Twenty-eight (51.8%) isolates tested in the study revealed resistance to all applied antibiotics except for polymyxin B.
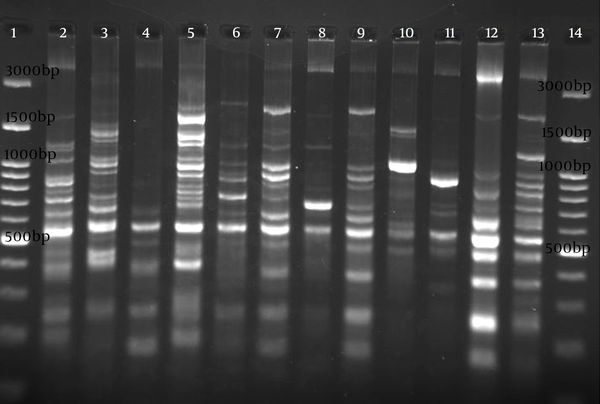
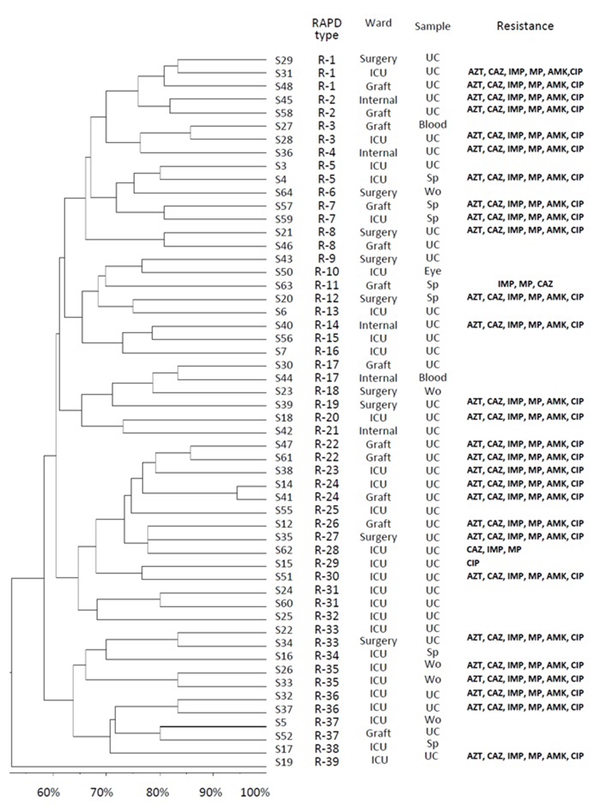
Electrophoresis patterns of RAPD-PCR for some isolates of studied P. aeruginosa are shown in Figure 2. Genetic relatedness of clinical isolates of the detected P. aeruginosa has been carried out using RAPD-PCR assay, which showed 55% to 100% similarity (Figure 3). The number of bands in RAPD typing varied from 2 to 12, with the length of 150 bp to 4300 bp. RAPD fingerprinting results of P. aeruginosa based on ≥ 80% showed 39 different groups (Figure 3). Although the majority of isolates had unique fingerprint, group one composed of 3 isolates (patients S48, S29, S31). These urine-derived isolates were from different wards. Isolate S29 was fully sensitive to applied antibiotics, whereas isolates S31 and S48 were resistance to applied antibiotics (Figure 3). Groups 22, 31, 35, and 36, each consisted of two isolates, were also obtained from the same ward (Figure 3). Different antibiotic resistance patterns were observed among the members of the same cluster. Based on 65% similarity level, eight main cluster , with most of the strains belonging to cluster 1 were detected (Figure 3).
Antibiotic Resistance Percentage of P. aeruginosa Isolated From Patients Admitted to Studied Hospital
Electrophoresis Image of RAPD Genotyping
Dendogram Showing Genetic Diversity Among 54 Non-duplicate Clinical Isolates of P. aeruginosa
5. Discussion
Constant monitoring of antibiotic susceptibility profiles and genetic relatedness among bacterial infectious agents are essential steps for infection control (34). To improve our understanding about P. aeruginosa antibiotic resistance and their distribution, 54 non-duplicate isolates of P. aeruginosa by using Kirby-Bauer’s disk diffusion and RAPD-PCR methods were investigated. P. aeruginosa involved in this study were highly resistant to different antibiotic families like, monobactams, cephalosporins, quinolones, and carbapenems (Figure 1). Our isolates had the highest resistance rate to imipenem and meropenem (55.6%), which is higher than similar reports from Iran (11-13). In addition, increased percentage of resistance to ciprofloxacin and amikacin (53.7% and 48.1%, respectively) were observed in comparison with previous study (29% and 17%) that have been performed in the same hospital (35). Although, in our investigated hospital, carbapenems (imipenem, meropenem) along with amikacin are the most currently used antibiotics for the treatment of P. aeruginosa infections, results of this study revealed that prescription of them should be limited.
In order to optimize treatment of P. aeruginosa infections in our hospital (also it can be mentioned as a limitation of this study), determination of resistance to other antibiotics like piperacillin, piperacillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, gentamicin, and cefepime, are essential to be evaluated in future studies. The finding of present study corresponds with the data presented by Haeili and associates, which showed the lowest percentage of resistance (0%) to polymyxin B (11).
There are reports that PFGE and MLST are more reliable methods for establishing clonal relatedness among P. aeruginosa strains, but because of the high cost, their utilization is limited, especially in developing countries (36, 37). Allegedly, RAPD typing is a valuable and useful method for the evaluation of genetic diversity among P. aeruginosa isolates (21, 23, 24). Out of various primers applied for RAPD-PCR, primer 272 was reliable because of its higher discriminatory power and reproducible profiles (38, 39). In the present study, all 54 isolates were typeable by RAPD-PCR; moreover, RAPD typing allowed us to reveal 54 unique finger prints among 54 clinical isolates of P. aeruginosa. Based on Dice similarity coefficient of greater than 80%, 39 groups were identified. Previous studies performed in Iran showed different distribution of genotypes. For instance, in the study of Salimi et al., by using RAPD typing, only 8 different groups among 129 isolates of P. aeruginosa were reported (40). In addition, in another study conducted by Nanvazadeh et al. 9 groups among 50 clinical samples of P. aeruginosa were observed (41). Using limited source for recovering P. aeruginosa may have limited the conclusions of these studies. General hygiene quality may also attribute to this genetic diversity. Although in a previous study the correlation between RAPD type and P. aeruginosa infection was suggested, our finding is not in agreement with these data (42).
Based on the level of 65% similarity, eight major clusters were observed. Isolates with shared antibiotic resistance profiles were placed in the separate groups. Furthermore, our data demonstrated a weak correlation between RAPD and antibiotic resistance profiles; it is supported by the results of other studies (34, 43). In a number of cases, isolates belonged to same cluster showed different antibiotic resistance patterns. The reason for this diversity may be attributed to the involved different antibiotic resistance mechanisms (5-7). Moreover; results of the independent studies showed that determination of antibiotic resistance pattern as phenotypic method for P. aeruginosa typing has low discriminatory power (34, 43). According to our data, the majority of isolates probably originated from host itself, but cross infection of P. aeruginosa is possible to occur in studied hospital. For instance, isolates S48, S29, and S31 were recovered from urine of the patients in different wards or isolates 26 and 33 from the same ward and sample had genetic similarity (based on RAPD fingerprinting results). Before taking any decision, we think that clonal relatedness of obtained isolates should be confirmed by more discriminatory methods, such as PFGE or MLST.
In conclusion, prescription of common antibiotics (imipenem and amikacin) in our hospital due to high resistance rate must be restricted. According to our data, considerable genetic diversity exists among isolated P. aeruginosa; it is possible that different sources of P. aeruginosa be involved in our hospital that can lead to host colonization. In order to accurately control the infection, other P. aeruginosa isolated from hospital environment must be subjected to fingerprinting. Our results did not reveal epidemic clone of P. aeruginosa.
Acknowledgements
References
-
1.
Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29(11):996-1011. [PubMed ID: 18947320]. https://doi.org/10.1086/591861.
-
2.
Rossolini GM, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11 Suppl 4:17-32. [PubMed ID: 15953020]. https://doi.org/10.1111/j.1469-0691.2005.01161.x.
-
3.
Gaynes R, Edwards JR, National Nosocomial Infections Surveillance S. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41(6):848-54. [PubMed ID: 16107985]. https://doi.org/10.1086/432803.
-
4.
Bouza E, Burillo A, Munoz P. Catheter-related infections: diagnosis and intravascular treatment. Clin Microbiol Infect. 2002;8(5):265-74. [PubMed ID: 12047403].
-
5.
Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10(4):441-51. https://doi.org/10.1586/erp.10.49.
-
6.
Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13(6):151-71. [PubMed ID: 20833577]. https://doi.org/10.1016/j.drup.2010.08.003.
-
7.
Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol. 2011;2.
-
8.
Pourakbari B, Sadr A, Ashtiani MT, Mamishi S, Dehghani M, Mahmoudi S, et al. Five-year evaluation of the antimicrobial susceptibility patterns of bacteria causing bloodstream infections in Iran. J Infect Dev Ctries. 2012;6(2):120-5. [PubMed ID: 22337839].
-
9.
Mohammadi-Mehr M, Feizabadi M. Antimicrobial resistance pattern of Gram-negative bacilli isolated from patients at ICUs of Army hospitals in Iran. Iran J Microbiol. 2011;3(1):26-30. [PubMed ID: 22347579].
-
10.
Hadadi A, Rasoulinejad M, Maleki Z, Yonesian M, Shirani A, Kourorian Z. Antimicrobial resistance pattern of Gram-negative bacilli of nosocomial origin at 2 university hospitals in Iran. Diagn Microbiol Infect Dis. 2008;60(3):301-5. [PubMed ID: 18036759]. https://doi.org/10.1016/j.diagmicrobio.2007.10.010.
-
11.
Haeili M, Ghodousi A, Nomanpour B, Omrani M, Feizabadi MM. Drug resistance patterns of bacteria isolated from patients with nosocomial pneumonia at Tehran hospitals during 2009-2011. J Infect Dev Ctries. 2013;7(4):312-7. [PubMed ID: 23592640]. https://doi.org/10.3855/jidc.2604.
-
12.
Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013;5(1):36-41. [PubMed ID: 23466812].
-
13.
Khosravi AD, Mihani F. Detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60(1):125-8. [PubMed ID: 17900848]. https://doi.org/10.1016/j.diagmicrobio.2007.08.003.
-
14.
Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006;19(3):512-30. https://doi.org/10.1128/CMR.00025-05.
-
15.
van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13 Suppl 3:1-46. [PubMed ID: 17716294]. https://doi.org/10.1111/j.1469-0691.2007.01786.x.
-
16.
Picard B, Denamur E, Barakat A, Elion J, Goullet P. Genetic heterogeneity of Pseudomonas aeruginosa clinical isolates revealed by esterase electrophoretic polymorphism and restriction fragment length polymorphism of the ribosomal RNA gene region. J Med Microbiol. 1994;40(5):313-22. [PubMed ID: 7909849].
-
17.
Römling U, Tümmler B. Achieving 100% Typeability of Pseudomonas aeruginosaby Pulsed-Field Gel Electrophoresis. J Clin Microbiol. 2000;38(1):464-5.
-
18.
Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42(12):5644-9. [PubMed ID: 15583294]. https://doi.org/10.1128/JCM.42.12.5644-5649.2004.
-
19.
Dawson SL, Fry JC, Dancer BN. A comparative evaluation of five typing techniques for determining the diversity of fluorescent pseudomonads. J Microbiol Methods. 2002;50(1):9-22. [PubMed ID: 11943354].
-
20.
Onteniente L, Brisse S, Tassios PT, Vergnaud G. Evaluation of the polymorphisms associated with tandem repeats for Pseudomonas aeruginosa strain typing. J Clin Microbiol. 2003;41(11):4991-7. https://doi.org/10.1128/JCM.41.11.4991-4997.
-
21.
Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34(5):1129-35. [PubMed ID: 8727889].
-
22.
Kidd TJ, Grimwood K, Ramsay KA, Rainey PB, Bell SC. Comparison of Three Molecular Techniques for Typing Pseudomonas aeruginosa Isolates in Sputum Samples from Patients with Cystic Fibrosis. J Clin Microbiol. 2010;49(1):263-8. https://doi.org/10.1128/jcm.01421-10.
-
23.
Campbell M, Mahenthiralingam E, Speert DP. Evaluation of random amplified polymorphic DNA typing of Pseudomonas aeruginosa. J Clin Microbiol. 2000;38(12):4614-5.
-
24.
Renders N, Romling Y, Verbrugh H, van Belkum A. Comparative typing of Pseudomonas aeruginosa by random amplification of polymorphic DNA or pulsed-field gel electrophoresis of DNA macrorestriction fragments. J Clin Microbiol. 1996;34(12):3190-5. [PubMed ID: 8940470].
-
25.
Mahon C, Lehman D, Manuselis G. Text Book of Diagnostic Microbiology. 4th ed. Elsevier; 2011.
-
26.
Tyler SD, Strathdee CA, Rozee KR, Johnson WM. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2(4):448-53. [PubMed ID: 7583922].
-
27.
Franklin R, Cockerill III. Performance standards for antimicrobial disk susceptibility tests Approved Standards, Twenty first informational supplements, M100- S21. Wayne, PA USA: Clinical laboratory standard Institute; 2011.
-
28.
Pavlicek A, Hrda S, Flegr J. Free-Tree--freeware program for construction of phylogenetic trees on the basis of distance data and bootstrap/jackknife analysis of the tree robustness. Application in the RAPD analysis of genus Frenkelia. Folia Biol (Praha). 1999;45(3):97-9. [PubMed ID: 10730897].
-
29.
Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357-8. [PubMed ID: 8902363].
-
30.
Ruimy R, Genauzeau E, Barnabe C, Beaulieu A, Tibayrenc M, Andremont A. Genetic diversity of Pseudomonas aeruginosa strains isolated from ventilated patients with nosocomial pneumonia, cancer patients with bacteremia, and environmental water. Infect Immun. 2001;69(1):584-8. [PubMed ID: 11119558]. https://doi.org/10.1128/IAI.69.1.584-588.2001.
-
31.
Giske CG, Libisch B, Colinon C, Scoulica E, Pagani L, Fuzi M, et al. Establishing clonal relationships between VIM-1-like metallo-beta-lactamase-producing Pseudomonas aeruginosa strains from four European countries by multilocus sequence typing. J Clin Microbiol. 2006;44(12):4309-15. [PubMed ID: 17021059]. https://doi.org/10.1128/JCM.00817-06.
-
32.
Libisch B, Balogh B, Fuzi M. Identification of two multidrug-resistant Pseudomonas aeruginosa clonal lineages with a countrywide distribution in Hungary. Curr Microbiol. 2009;58(2):111-6. [PubMed ID: 18946702]. https://doi.org/10.1007/s00284-008-9285-7.
-
33.
Libisch B, Watine J, Balogh B, Gacs M, Muzslay M, Szabo G, et al. Molecular typing indicates an important role for two international clonal complexes in dissemination of VIM-producing Pseudomonas aeruginosa clinical isolates in Hungary. Res Microbiol. 2008;159(3):162-8. [PubMed ID: 18280707]. https://doi.org/10.1016/j.resmic.2007.12.008.
-
34.
Czekajlo-Kolodziej U, Giedrys-Kalemba S, Medrala D. Phenotypic and genotypic characteristics of Pseudomonas aeruginosa strains isolated from hospitals in the north-west region of Poland. Pol J Microbiol. 2006;55(2):103-12. [PubMed ID: 17419287].
-
35.
Fazeli H, Akbari R, Moghim S, Narimani T, Arabestani MR, Ghoddousi AR. Pseudomonas aeruginosa infections in patients, hospital means, and personnel's specimens. J Res Med Sci. 2012;17(4):332-7. [PubMed ID: 23267393].
-
36.
Johnson JK, Arduino SM, Stine OC, Johnson JA, Harris AD. Multilocus sequence typing compared to pulsed-field gel electrophoresis for molecular typing of Pseudomonas aeruginosa. J Clin Microbiol. 2007;45(11):3707-12. [PubMed ID: 17881548]. https://doi.org/10.1128/JCM.00560-07.
-
37.
Waters V, Zlosnik JE, Yau YC, Speert DP, Aaron SD, Guttman DS. Comparison of three typing methods for Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Eur J Clin Microbiol Infect Dis. 2012;31(12):3341-50. [PubMed ID: 22843295]. https://doi.org/10.1007/s10096-012-1701-z.
-
38.
Saitou K, Furuhata K, Fukuyama M. Genotyping of Pseudomonas aeruginosa isolated from cockroaches and human urine. J Infect Chemother. 2010;16(5):317-21. [PubMed ID: 20352277]. https://doi.org/10.1007/s10156-010-0055-7.
-
39.
Lanotte P, Watt S, Mereghetti L, Dartiguelongue N, Rastegar-Lari A, Goudeau A, et al. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol. 2004;53(Pt 1):73-81. [PubMed ID: 14663109].
-
40.
Salimi H, Owlia P, Yakhchali B, Lari AR. Drug Susceptibility and Molecular Epidemiology of Pseudomonas aeruginosa Isolated in a Burn Unit. Am J Infect Dis. 2009;5(4):301-6.
-
41.
Nanvazadeh F, Khosravi AD, Zolfaghari MR, Parhizgari N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns. 2013;39(7):1409-13. [PubMed ID: 23773789]. https://doi.org/10.1016/j.burns.2013.03.008.
-
42.
Matar GM, Harakeh HS, Ramlawi F, Khneisser I, Hadi U. Comparative analysis between Pseudomonas aeruginosa genotypes and severity of symptoms in patients with unilateral or bilateral otitis externa. Curr Microbiol. 2001;42(3):190-3. [PubMed ID: 11270653].
-
43.
Nazik H, Ongen B, Erturan Z, Salcioglu M. Genotype and antibiotic susceptibility patterns of Pseudomonas aeruginosa and Stenotrophomonas maltophilia isolated from cystic fibrosis patients. Jpn J Infect Dis. 2007;60(2-3):82-6. [PubMed ID: 17515637].