Abstract
Background:
Urinary tract infections (UTIs) are one of main health problems caused by many microorganisms, including uropathogenic Escherichia coli (UPEC). UPEC strains are the most frequent pathogens responsible for 85% and 50% of community and hospital acquired UTIs, respectively. UPEC strains have special virulence factors, including type 1 fimbriae, which can result in worsening of UTIs.Objectives:
This study was performed to detect type 1 fimbriae (the FimH gene) among UPEC strains by molecular method.Materials and Methods:
A total of 140 isolated E. coli strains from patients with UTI were identified using biochemical tests and then evaluated for the FimH gene by polymerase chain reaction (PCR) analysis.Results:
The UPEC isolates were identified using biochemical tests and were screened by PCR. The fimH gene was amplified using specific primers and showed a band about 164 bp. The FimH gene was found in 130 isolates (92.8%) of the UPEC strains. Of 130 isolates positive for the FimH gene, 62 (47.7%) and 68 (52.3%) belonged to hospitalized patients and outpatients, respectively.Conclusions:
The results of this study indicated that more than 90% of E. coli isolates harbored the FimH gene. The high binding ability of FimH could result in the increased pathogenicity of E. coli; thus, FimH could be used as a possible diagnostic marker and/or vaccine candidate.Keywords
Urinary Tract Infections Uropathogenic Escherichia Coli FimH
1. Background
Urinary tract infections (UTIs) are one of the inflammatory diseases produced by high multiplication of many pathogens in the urinary apparatus, resulting in alterations in the perfect function of the urinary tract and kidneys. UTI is particularly a major problem for females; nearly 50–80% of the female population endures from UTI at least once in lifetime and 20-50% of them will have recrudescent events (1, 2).
Escherichia coli is the most frequent pathogen responsible for up to 80% of UTIs (3). This bacteria is responsible for 85% and 50% of community and hospital acquired UTIs, respectively (4). Uropathogenic E. coli (UPEC) strains have special virulence factors, including pili or fimbriae, which mediate attachment to uroepithelial and vaginal cells, resistance to human serum bactericidal activity, haemolysin production, and increased amounts of K capsular antigen (5). Furthermore, virulence factors of UPEC strains have a significant role in development of UTIs. The most virulence factors dependent upon the UPEC include adhesions (type 1 fimbriae, p fimbriae, curli fimbriae, afimbrial adhesion and flagellum), aerobactins, hemolysins, and cytotoxic necrotizing factor 1. The mentioned virulence factors are important in colonization of UPEC, extra-intestinal survival, and creation of cytopathic effects. In addition, the expression of special virulence factors of UPEC can contribute to uropathogenicity, as well as worsening of UTIs (1, 6, 7).
An essential step for beginning and development of UTI is bacterial attachment to uroepithelial cells. E. coli attachment is mediated by ligands of bacteria (generally small proteins placed at the tips of bacterial fimbriae) which bind to host cell wall carbohydrate residues, working as receptors (5). Therefore, the adherence of E. coli to host receptors is a function, usually mediated by adhesions of bacteria to host cell receptors (8). The bacterial attachment permits bacteria to resist mechanical elimination by the flow of urine and bladder emptying and increasing persistence of E. coli. UPEC strains produce different types of adhesins, including type 1 fimbriae, which are essential for recognition and attachment to urinary tract receptors (9).
Among adhesions of UPEC, the adhesive subunit of type 1 fimbriae, FimH, is a major determinant, which has high tropism for urinary tract receptors; thus, FimH adhesion is important in colonizing different niches of E. coli (10). In addition, single-nucleotide polymorphism (SNP) analysis of FimH is a screening tool for epidemiological typing of UPEC (11, 12). Therefore, the research on bacterial virulence factors can result in expansion and development of new methods for diagnosis and prevention of UTIs. For more subsequent investigations, the fimH gene was detected in UPEC strains isolated from hospitalized and out-patients with UTI, referred to educational hospitals of Shahrekord.
2. Objectives
The present study was conducted for detection of the fimH virulence gene from UPEC isolated from both hospitalized patients and outpatients with UTI, referred to educational hospitals of Shahrekord, Iran.
3. Materials and Methods
3.1. Bacterial Isolates
In this study, 140 isolated E. coli strains from patients with UTI were evaluated. The isolates were collected from both hospitalized and non-hospitalized patients with UTIs from April to July 2012 from Kashani and Hajar hospitals, Shahrekord, Iran.
Hospitalized infections were characterized as patients who were confined to bed in hospital and non-hospitalized infections were characterized as infections in patients who had no prior contacts with hospitals or long-term care facilities two weeks prior to admission.
The bacterial isolates were identified using biochemical tests. The samples were cultured on MacConkey agar, blood agar and eosin methylene blue (EMB) agar (Hi Media, India). The plates were incubated at 35°C for 24 hours and pure isolates were characterized and identified according to Gram staining and biochemical tests such as catalase, oxidase, indole production, citrate utilization, triple iron sugar, ortho-nitrophenyl-β-galactoside (ONPG) test, and methyl red-Voges Proskauer, as described in standard bacteriological methods. All the above chemicals and media were purchased from Sigma-Aldrich (Germany).
3.2. DNA Extraction for Polymerase Chain Reaction
Genomic DNA templates for PCR amplification were gained from overnight growth of bacterial isolates on Luria-Bertani agar (Hi Media, India), pelleted by centrifugation, resuspended in 500 µL of sterile deionized water, and boiled for 10 minutes. After centrifugation of the boiled samples at 19000 g for five minutes, the supernatant was applied as the DNA template for PCR.
3.3. Polymerase Chain Reaction Amplification of β-Lactamase Genes
The bacterial isolates were subjected to screening for the presence of the fimH gene by PCR. The nucleotide sequence of the primers and the annealing temperature for amplification of the fimH gene by PCR are shown in Table 1. PCR was carried out in 25 µL containing 2.5 µL of 10 × PCR reaction buffer with MgCl2 (1.6 mm), 0.5 µL (200 µM) of deoxynucleoside triphosphates mixture (dNTPs, 10 mm), 0.5 µL of each primer (10 pm/µL), 2 µL of the DNA template (50 ng) with 0.5 µL (3 U/µL) Taq DNA polymerase. The amplification condition included an initial denaturation (96°C for three minutes), 37 cycles (96°C for 30 seconds, 64°C for 30 seconds, 72°C for 60 seconds) and a final extension (72°C for fiv eminutes). The PCR amplifications were performed on a Thermocycler TC-512 (Bio-Techne, Cambridge UK) and the PCR products were analyzed by polyacrylamide gel (8%) electrophoresis. A molecular marker (Fermentas SM0321, 100 bp) was used to assess the PCR products sizes. The PCR materials were purchased from Fermentas (Life Science, Germany).
Primers and Annealing Temperature for Amplification of the fimH Gene
| Primers | Temperature, °C | Nucleotide Sequences (5´-3´) | Size, bp |
|---|---|---|---|
| FimH-F | 64 | GTGCCAATTCCTCTTACCGTT | 164 |
| FimH-R | TGGAATAATCGTACCGTTGCG |
4. Results
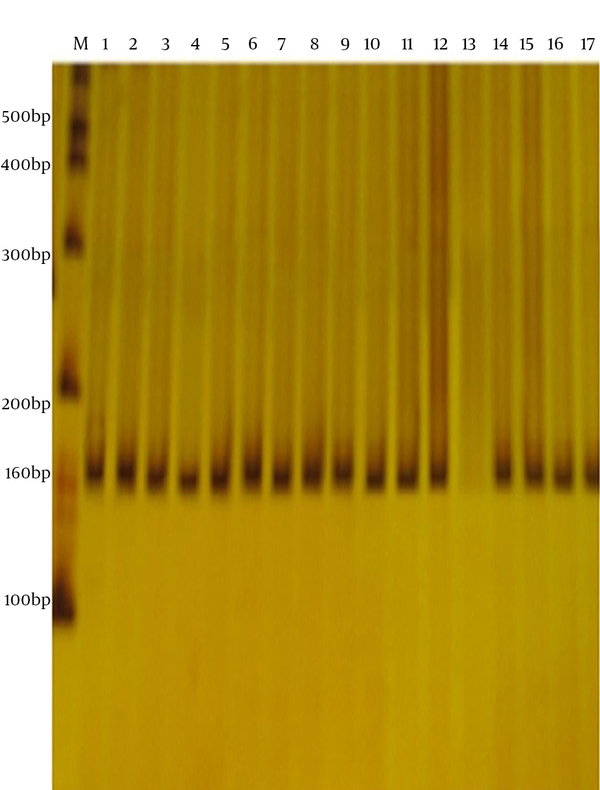
All the bacterial isolates were identified using biochemical tests and screened by PCR. The fimH gene was amplified using specific primers and appeared as a band of about 164 bp on polyacrylamide gel (Figure 1). The FimH gene was found in 130 isolates (92.8%) of UPEC. Of the 130 isolates positive for the FimH gene, 62 (47.7%) and 68 (52.3%) belonged to hospitalized patients and outpatients, respectively.
Polymerase Chain Reaction Amplification of the FimH Gene
5. Discussion
The onset of UTI results from the ability of UPEC attachment to urinary tract epithelial cells by specific adhesions including type 1 fimbriae (13). UPEC strains have many virulence factors which enhance their capacity to colonize in the urogenital tract. Attachment to the urothelial cell surface is mediated by FimH adhesion, placed at the tip of the type 1 fimbriae, which prevents bacterial washout by urine flow and starts bacterial invasion (4, 14). Since the FimH virulence factor associated with UTI cases was not widely determined from the UPEC isolated from both hospitalized patients and outpatients with UTIs referred to educational hospitals of Shahrekord, Iran, the prevalence of the fimH gene was examined.
The presence of the fimH gene was confirmed by PCR and the results indicated that the fimH gene was present in 130 UPEC isolates (92.8%); 62 isolates (47.7%) of hospitalized patients and 68 (52.3%) of outpatients. This showed that most of the UPEC strains had the fimH gene and our results were almost in accordance with the results of previous literature. Tarchouna et al. (15) reported that among the studied virulence genes of UPEC strains, the fimH gene was the most prevalent virulence gene and was found in 68% (61/90) of the UTI isolates. Garofalo et al. (16) studied 18 UPEC isolates collected from females and found that the fimH gene was the most prevalent virulence factor and 100% of the isolates had that gene. In another study, Watts et al. (17) demonstrated that the fimH gene was the most frequent virulence gene and was detected in 98% of E. coli strains isolated from patient with UTIs. Mladin et al. (7) evaluated the distribution of virulence genes among the studied UPEC and showed that the prevalence of different virulence genes varied from 10% for the cnf gene to 80% for the fimH gene. In addition, Arabi et al. (18) investigated the frequency of FimH and other adhesions genes in UPEC and determined the fimH gene frequency as 87.7%.
Apart from investigation on evaluation of the FimH gene in UPEC strains, this gene has been detected in other strains of E. coli. For example, Kaczmarek et al. (13) evaluated and detected the genes encoding virulence factors among E. coli strains with K1 antigen as well as the non-K1 E. coli strains. They found that the FimH gene existed in the whole tested E. coli K1strains as well as in 97.0% of non-K1 strains. Catana et al. (19) also reported that the fimH gene was present in 54.55% of avian pathogenic E. coli (APEC) strains which are pathogenic for birds. In another study, Biscola et al. (20) showed that in enterohemorrhagic E. coli (EHEC) strains, the fimH gene was conveyed by the majority of non-O157:H7 E. coli strains (97%) and by all the O157:H7 E. coli strains. Fernandes et al. (21) detected the FimH gene in all the E. coli isolates from bovine mastitis. Moreover, FimH SNP analysis is a typing tool for epidemiological studying of community- and hospital-associated E. coli isolates and could be used as an easy, cheap screening test for genotypic analyses of UPEC (11, 12).
We can conclude that type 1 fimbriae is present among UPEC as well as other strains of E. coli, to the extent that the FimH gene was detected in more than 90% of the E. coli strains. The high binding ability of FimH could result in increased bacterial binding to target cells and increased pathogenicity of E. coli; thus, FimH could be used to design vaccine for prevention of E. coli infections by blocking the bacterial attachment and colonization. In addition, FimH could be used as a tool for the extension of rapid detection-based assays.
Acknowledgements
References
-
1.
Agarwal J, Srivastava S, Singh M. Pathogenomics of uropathogenic Escherichia coli. Indian J Med Microbiol. 2012;30(2):141-9. [PubMed ID: 22664427]. https://doi.org/10.4103/0255-0857.96657.
-
2.
Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113 Suppl 1A:5S-13S. [PubMed ID: 12113866].
-
3.
Wojnicz D. Virulence factors of uropathogenic Escherichia coli strains isolated from children with chronic pyelonephritis. Adv Clin Exp Med. 2007;16(5):651.
-
4.
Su C. Female lower urinary tract infection. JTUA. 2008;19:12-20.
-
5.
Emody L, Kerenyi M, Nagy G. Virulence factors of uropathogenic Escherichia coli. Int J Antimicrob Agents. 2003;22 Suppl 2:29-33. [PubMed ID: 14527768].
-
6.
Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27(4):505-23. [PubMed ID: 14550943].
-
7.
Mladin C, Usein CR, Chifiriuc M, Palade A, Slavu CL, Negut M, et al. Genetic analysis of virulence and pathogenicity features of uropathogenic Escherichia coli isolated from patients with neurogenic bladder. Rom Biotech Lett. 2009;14(6):4906-11.
-
8.
Le Bouguenec C. Adhesins and invasins of pathogenic Escherichia coli. Int J Med Microbiol. 2005;295(6-7):471-8. [PubMed ID: 16238021].
-
9.
Oliveira FA, Paludo KS, Arend LN, Farah SM, Pedrosa FO, Souza EM, et al. Virulence characteristics and antimicrobial susceptibility of uropathogenic Escherichia coli strains. Genet Mol Res. 2011;10(4):4114-25. [PubMed ID: 22057993]. https://doi.org/10.4238/2011.October.31.5.
-
10.
Sokurenko EV, Feldgarden M, Trintchina E, Weissman SJ, Avagyan S, Chattopadhyay S, et al. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol Biol Evol. 2004;21(7):1373-83. [PubMed ID: 15044596]. https://doi.org/10.1093/molbev/msh136.
-
11.
Dias RC, Moreira BM, Riley LW. Use of fimH single-nucleotide polymorphisms for strain typing of clinical isolates of Escherichia coli for epidemiologic investigation. J Clin Microbiol. 2010;48(2):483-8. [PubMed ID: 20018817]. https://doi.org/10.1128/JCM.01858-09.
-
12.
Tartof SY, Solberg OD, Riley LW. Genotypic analyses of uropathogenic Escherichia coli based on fimH single nucleotide polymorphisms (SNPs). J Med Microbiol. 2007;56(Pt 10):1363-9. [PubMed ID: 17893175]. https://doi.org/10.1099/jmm.0.47262-0.
-
13.
Kaczmarek A, Budzynska A, Gospodarek E. Prevalence of genes encoding virulence factors among Escherichia coli with K1 antigen and non-K1 E. coli strains. J Med Microbiol. 2012;61(Pt 10):1360-5. [PubMed ID: 22745135]. https://doi.org/10.1099/jmm.0.044263-0.
-
14.
Finer G, Landau D. Pathogenesis of urinary tract infections with normal female anatomy. Lancet Infec Dis. 2004;4(10):631-5.
-
15.
Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013;17(6):e450-3. [PubMed ID: 23510539]. https://doi.org/10.1016/j.ijid.2013.01.025.
-
16.
Garofalo CK, Hooton TM, Martin SM, Stamm WE, Palermo JJ, Gordon JI, et al. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007;75(1):52-60. [PubMed ID: 17074856]. https://doi.org/10.1128/IAI.01123-06.
-
17.
Watts RE, Hancock V, Ong CL, Vejborg RM, Mabbett AN, Totsika M, et al. Escherichia coli isolates causing asymptomatic bacteriuria in catheterized and noncatheterized individuals possess similar virulence properties. J Clin Microbiol. 2010;48(7):2449-58. [PubMed ID: 20444967]. https://doi.org/10.1128/JCM.01611-09.
-
18.
Arabi S, Tohidi F, Naderi S, Nazemi A, Jafarpour M, Naghshbandi R. The common fimbarie genotyping in Uropathogenic Escherichia coli. Ann Biol Res. 2012;3(10):4951-4.
-
19.
Catana N, Popa V, Fodor I. Binding of Congo Red, Phenotypical Marker for Discrimination of Apec Strains. Lucrari Stiintifice-Universitatea de Stiinte Agricole a Banatului Timisoara, Medicina Veterinara. 2009;42(1):200-2.
-
20.
Biscola FT, Abe CM, Guth BE. Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol. 2011;77(7):2201-8. [PubMed ID: 21317257]. https://doi.org/10.1128/AEM.01920-10.
-
21.
Fernandes JB, Zanardo LG, Galvao NN, Carvalho IA, Nero LA, Moreira MA. Escherichia coli from clinical mastitis: serotypes and virulence factors. J Vet Diagn Invest. 2011;23(6):1146-52. [PubMed ID: 22362795]. https://doi.org/10.1177/1040638711425581.