Abstract
Background:
Escherichia coli can cause human diseases and cow mastitis. Use of antibiotics in the treatment of E. coli infections causes drug residues in dairy products and develops antibiotic resistance. Thus, it is of prime importance to find a new solution for the treatment of E. coli infection.Objectives:
The present study aimed at constructing a novel cell epitope-based polypeptide vaccine (OmpC-EP) against E. coli infection.Methods:
Based on the outer membrane protein C (OmpC) of E. coli, we used ABCpred and BepiPred method to obtain its B cell epitopes, whereas nHLAPred and ProPred methods were used to obtain the CTL and Th cell epitopes, respectively. The predicted cell epitopes were recomposed using DNASTAR software to obtain a cell epitope-based polypeptide (OmpC-EP) of high antigenicity, which was purified by Ni-NTA flow resin. Purified OmpC-EP was mice immunized to prepare a polyclonal antibody; Western blotting analysis was used to detect antibody specificity. Pull down and ELISA detected the interaction between OmpC-EP antibodies and E. coli. Active immunity mice and challenge of E. coli were used to detect the immune protection of CM-TEP.Results:
A novel epitope peptide (OmpC-EP), with a MW of 12.5 kDa was designed, and Ni-NTA purified OmpC-EP was mice immunized to prepare a polyclonal antibody. OmpC-EP antiserum had a good specificity and directly interacted with E. coli. Specific immunity was activated in mice, and OmpC-EP displayed a significant immune protective effect (62.5%), which was slightly higher than that of the OmpC protein (56.3%) following infection with E. coli.Conclusions:
The results of the present study revealed that OmpC-EP has a good immunogenicity and possesses a significant immune protective function. OmpC-EP is expected to be an approach to construct a vaccine against E. coli.Keywords
OmpC Protein Cell Epitope Immune Protection Escherichia coli
1. Background
Escherichia coli is a Gram-negative bacterium, facultative anaerobic, rod-shaped, and coliform bacterium of the genus Escherichia. Pathogenic E. coli strains can be categorized based on elements that can elicit an immune response in animals including O antigen, K antigen, and H antigen. E. coli can enter the digestive tract through the mouth, causing serious food poisoning, diarrhea, sepsis (1), and meningitis in children (2). Escherichia coli can also cause mastitis in cows (3), which can seriously affect the quality of dairy products, leading to huge economic losses to the dairy industry every year. Thus, E. coli is an important zoonotic pathogen (4).
Currently, use of antibiotics in the treatment of E. coli infections causes drug residues in dairy products, and develops antibiotic resistance (5, 6).Thus, it is necessary to find a new solution for the treatment of E. coli infections. The E. coli outer membrane protein (OmpC protein), which is known to stimulate humoral and cellular immunity in animals, is an important candidate for constructing a vaccine (7, 8). To improve the immune function of the OmpC protein, we constructed an OmpC epitope vaccine.
Epitope vaccines are a new type of vaccines developed in recent years. Compared to traditional vaccines, epitope vaccines can easily be recognized by major histocompatibility complex (MHC) molecules with different genetic backgrounds (9); they have also been studied for use against viruses (10), bacteria (11), and cancer immunotherapy (12). However, epitope peptide vaccines have rarely been investigated for the treatment of E. coli infections.
2. Objectives
Based on cell epitope vaccine, this study was aimed at designing and constructing a novel cell epitope-based polypeptide vaccine (OmpC-EP) against E. coli infection, and verifying its immunological function, laying a foundation for the construction of an OmpC protein epitope vaccine.
3. Methods
3.1. Ethics Statement
This study was approved by the animal ethical committee (Ref. no20150207), Shaanxi University of Technology, China.
3.2. Standard Materials
The E. coli OmpC amino acid sequence was obtained from the NCBI GenBank (accession number: ADU34074.2). Synthesis and cloning of the nucleic acid sequences of the epitope peptide were performed by the Shanghai Xuguan Biotech Development Corp., China. E. coli intestinal pathogenic bacteria Y17 was obtained from Shaanxi University of Technology, China. Kunming mice were purchased from Xian Jiaotong University College of Medicine, China.
3.3. Cell Epitope Analysis
The BepiPred 1.0b (http://www.cbs.dtu.dk/services/ BepiPred/) and ABCpred (http://www.imtech. res. in/raghava/abcpred/ABC submission. html) (13) software programs were used to predict the B cell epitopes of the E. coli OmpC protein. The neural network–based MHC Class-I Binding Peptide Prediction Server (nHLAPred) (http://www.imtech.res.in/raghava/nhlapred/comp.html) (10) and the MHC Class-II Binding Peptide Prediction Server (ProPred) (http://www.imtech.res.in/raghava/propred/) (11) were used to predict the cell epitope peptide segments of cytotoxic T lymphocytes (CTL) and helper T cells (Th), respectively. The DNASTAR software was used to optimize the arrangement of the cell epitopes as the final OmpC epitope peptide (OmpC-EP).
3.4. Prokaryotic Expression of OmpC-EP
OmpC-EP nucleic acid sequences were synthesized and cloned in E. coli BL21 by the company. E. coli BL21 harboring the expression vector was induced by adding 0.1 mM isopropyl-β-D-thiogalactoside (IPTG) (Sigma, USA). Bacterial cells were harvested by centrifugation after 5 hours following IPTG addition. All samples were heated for 5 minutes in boiling water and electrophoresed by SDS-PAGE. The protein bands were visualized by staining with G-250 (Sigma, USA) (14).
3.5. Purification of OmpC-EP
Bacterial cells were harvested from a large number of cultures and resuspended in 50 mM sodium phosphate buffer (pH 8.0) containing 8 M urea and were disrupted by sonication in an ice bath. The supernatant was collected and loaded into a column packed with Ni2+-nitriloaceate and purified by affinity chromatography on Ni-NTA flow resin (Sigma, USA) (15).
3.6. OmpC-EP Antisera Preparation
Five-week-old Kunming mice with purified protein (100 μg per mouse) were used and emulsified with Freund’s Complete Adjuvant (Shanghai Sangon Biotech. Corp., China), followed by other 2 injections with Freund’s Incomplete Adjuvant (Shanghai Sangon Biotech. Corp., China) at intervals of 14 days. Serum samples were collected from the mice and stored at -80°C.
3.7. Western Blotting Analysis for OmpC-EP Antisera Specificity
Western blotting was performed as described previously (14). Escherichia coli was prepared for SDS-PAGE and transferred into a nitrocellulose (NC) membrane (Sigma, USA). Once the transfer was complete, the membrane was removed and incubated in blocking solution (5% skim milk). Then, the NC membrane was incubated with various dilutions of mouse anti-OmpC-EP for 1 hour at 37°C. After rinsing, the NC membrane was incubated with rabbit anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (Sigma, USA). The membranes were developed with the dim-ethylaminoazobenzene (DAB) (Shanghai Sangon Biotech. Corp., China) substrate system until maximum bands visualization was reached.
3.8. Pull down and ELISA Assays to Detect the Interaction Between OmpC-EP Antiserum and E. coli
The bacterial pull down assay was performed as described previously (14). Briefly, E. coli cultures of OD600 1.0 were harvested and washed with physiological saline (0.85% NaCl). After addition of 1% oxymethylene (W/V) for 90 minutes at 80°C and centrifugation at 8000 × g for 10 minutes, the bacterial pellet was resuspended in physiological saline and the final concentration was adjusted to 0.2 OD at 600 nm. One milliliter of bacterial suspension was transferred into each 1.5 mL tube. After centrifugation, 100 μL OmpC-EP antiserum at various dilutions was added to the tubes and 2 μg/μL BSA was used as the negative control. Then, the bacteria were incubated with rabbit anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies. After suspending the bacteria in 20 μL PBS, the samples were transferred to enzyme-labeled plates, and coloration liquid (50 μL H2O2 and 50 μL TMB) was added to each well. After the color reaction, 50 μL stop solution (2 M H2SO4) was added to each well and the absorbance was read at 450 nm using a microplate reader (Bio-Rad, USA).
3.9. Active Immunity and Challenge
Mice were randomly divided into groups, and purified OmpC-EP protein was intraperitoneally injected 2 times at an interval of 10 days. OmpC-EP protein was emulsified with Freund’s complete adjuvant (100 μg per mouse) and Freund’s incomplete adjuvant (100 μg per mouse) was utilized in the primary and booster immunizations, respectively. Immunized mice were intraperitoneally challenged with 1.0 × 108 CFU/mL E. coli intestinal pathogenic bacterial strain Y17 (15). The control groups were injected with OmpC protein, PBS, and Freund’s adjuvant, respectively. Mice were observed for 15 days to measure their relative percent survivals (RPS). Protection rates were calculated using the following formula: RPS (%) = 1 - (% vaccinated mortality/% non-vaccinated mortality) × 100. Statistical significance between groups was tested using SPSS software (15).
4. Results
4.1. In Silico Prediction of B Cell Epitopes
The B cell epitope of the E. coli OmpC protein was predicted using BepiPred 1.0b (Table 1) and ABCpred (Table 2). Based on the common epitope sequence predicted by the 2 programs at residues 74 - 90, 129 - 144, 146 - 152, 178 - 194, 213 - 227, the B cell epitope segment of the OmpC protein was confirmed.
Identification of B Cell Epitopes for the E. coli OmpC Protein Using the BepiPred 1.0b Method
| Prediction Method | Peptide Segment Position | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| BepiPred | 46 - 55 | 64 - 93 | 129 - 144 | 146 - 152 | 171 - 203 | 213 - 244 | 256 - 262 | 269 - 275 | 346 - 354 |
Identification of B Cell Epitopes for the E. coli OmpC Protein Using the ABCpred Method
| Analysis Index | Analysis Results | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peptide segment position | 129 - 145 | 275 - 291 | 302 - 318 | 178 - 194 | 140 - 156 | 120 - 136 | 211 - 227 | 74 - 90 | 318 - 334 |
| Score | 0.95 | 0.93 | 0.92 | 0.92 | 0.92 | 0.92 | 0.91 | 0.90 | 0.90 |
4.2. In Silico Prediction of CTL Cell Epitopes
For nHLAPred-based CTL cell epitope prediction, we used HLA-A2, HLA-A*0201, HLA-A*0202, HLA-A*0203, and HLA-A*0205. Based on the common epitope sequences, the CTL cell epitope segment of the OmpC protein was identified as GLRPSLAYL, corresponding to positions 291 to 299 of OmpC (Table 3).
Prediction of CTL Epitopes for the E. coli OmpC Protein
| Allele | Position | Sequence |
|---|---|---|
| HLA-A2 | 5 - 15 | VLSLLVPALLV |
| 73 - 81 | GYGQWEYQI | |
| 113 - 121 | RNYGVVYDV | |
| 253 - 262 | IYLAAQYTQT | |
| 291 - 299 | GLRPSLAYL | |
| HLA-A*0201 | 5 - 15 | VLSLLVPALLV |
| 291 - 299 | GLRPSLAYL | |
| HLA-A*0202 | 7 - 15 | SLLVPALLV |
| 193 - 201 | ALRQNGDGV | |
| 291 - 299 | GLRPSLAYL | |
| HLA-A*0203 | 254 - 262 | YLAAQYTQT |
| 284 - 292 | AQYQFDFGL | |
| HLA-A*0205 | 5 - 17 | VLSLLVPALLVAG |
| 33 - 41 | DLYGKVDGL | |
| 91 - 105 | NSWTRVAFAGLKFQD | |
| 156 - 164 | DFFGLVDGL | |
| 186 - 194 | VTNNGRDAL | |
| 226 - 239 | AQNTAAYIGNGDRA | |
| 288 - 299 | FDFGLRPSLAYL | |
| 314 - 322 | DILKYVDVG |
4.3. In Silico Prediction of Th Cell Epitopes
Using the ProPred method and analyzing 3 different types of peptides, DRB1-0101, DRB1-0102, and DRB1-0301, we predicted the Th epitopes of E. coli OmpC (Table 4). This analysis revealed that the FGIGGAISS segment of OmpC, corresponding to residues 212 to 220, represents the Th cell epitope.
Prediction of Th Epitopes for the E. coli OmpC Protein
| Type | Peptide Segment Position | Sequence |
|---|---|---|
| DRB1-0101 | 5 - 22 | VLSLLVPALLVAGAANAA |
| 110 - 119 | YGRNYGVVY | |
| 141 - 149 | FMQQRGNGF | |
| 152 - 166 | YRNTDFFGLVDGLNF | |
| 212 - 220 | FGIGGAISS | |
| 298 - 306 | YLQSKGKNL | |
| 325 - 335 | YYFNKNMSTYV | |
| DRB1-0102 | 5 - 22 | VLSLLVPALLVAGAANAA |
| 158 - 166 | FGLVDGLNF | |
| 212 - 220 | FGIGGAISS | |
| 292 - 300 | LRPSLAYLQ | |
| DRB1-0301 | 5 - 17 | VLSLLVPALLVAG |
| 57 - 65 | MRLGFKGET | |
| 100 - 108 | LKFQDVGS | |
| 117 - 125 | VVYDVTSWT | |
| 212 - 220 | FGIGGAISS | |
| 246 - 254 | LKYDANNIY | |
| 352 - 367 | YYFNKNMSTYV |
4.4. Reorganization of the Epitope Series
The B, CTL, and Th epitopes were numbered, and 4 glycine residues (GGGG) were added to the epitope to increase the flexibility of the peptide. Then, different possible arrangements and combinations were optimized using the DNAStar software (Figure 1), and the final amino acid sequence was as follows:
AISSSKRTDAQNTAAGGGGYDVTSWTDVLPEFGGDGGGGV QYQGKNGSVSGEGMTNGGGGFGIGGAISSGGGGGLRPSLAYLGGGGETQVTDQLTGYGQWEYQGGGGYGSDNFM; the flexible portions are underlined.
Antigenic Index of the OmpC-EP

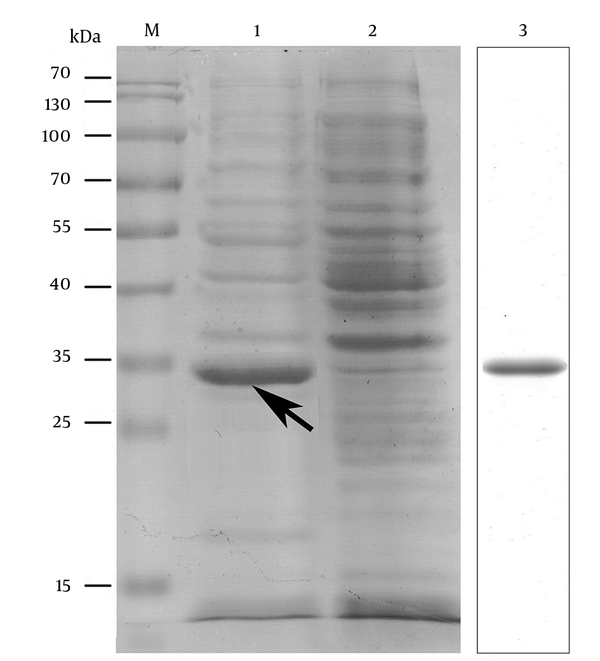
4.5. Prokaryotic Expression, Purification, and Antibody Preparation of OmpC-EP
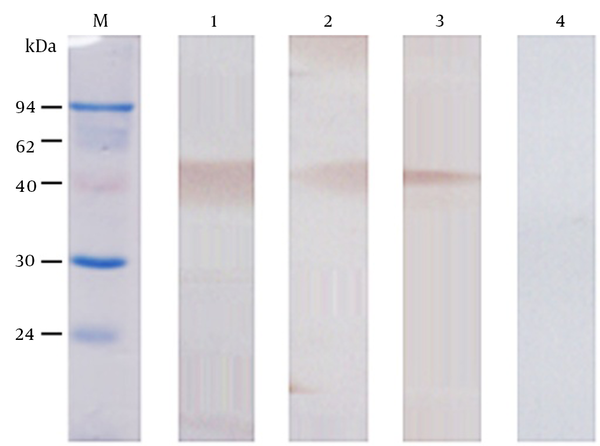
The nucleic acid sequence of the designed OmpC-EP peptide was sent to the company and cloned into a pET-32a plasmid and transformed into an expression strain of E. coli BL21. Heterologous expression of OmpC-EP in E. coli and induction with IPTG resulted in the production of a protein of about 33 kDa in size, as revealed by SDS-PAGE electrophoresis. This is the expected size of the recombinant protein, which corresponds to a fusion protein of 20.4 kDa and OmpC-EP of 12.5 kDa (Figure 2). To obtain a reasonable quantity of recombinant protein for subsequent analyses, Ni-NTA super flow resin was used (Figures 2 and 3). The purified recombinant protein was used for antibody preparation, and the specificity of the prepared antibody was validated using Western blot analysis. Only a single band was stained at the molecular mass of the corresponding protein. Moreover, the band intensity decreased gradually as the antibody concentration was reduced, indicating that OmpC-EP antiserum had a good specificity (Figure 3).
Expression and Purification of OmpC-EP
Determination of the Specificity of OmpC-EP Antiserum by Western Blot Analysis
4.6. Interaction Between OmpC-EP Peptide Antiserum and E. coli
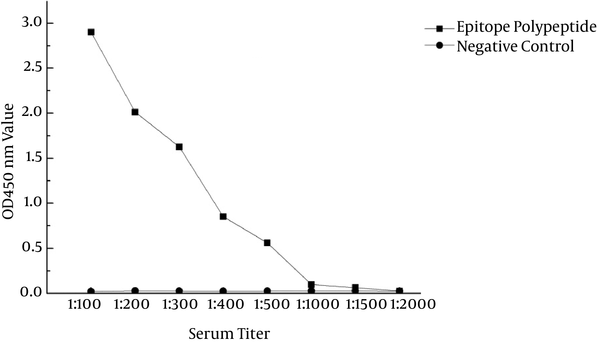
ELISA was used to determine the relationship between E. coli and OmpC-EP antibodies. The OD at 450 nm decreased with as serum dilution increased, and it was almost zero in control serum (Figure 4). These data indicated a direct interaction between OmpC-EP antiserum and E. coli, and showed good immunogenicity.
The Interaction Between OmpC-EP Antiserum and E. coli
4.7. Immunization of Mice with OmpC-EP Protects Against E. coli Infection
An active immunization approach was used to investigate the ability of OmpC-EP to protect mice against infection. Following infection with E. coli, mice showed severe symptoms including fluffy hair, wrinkled skin, decreased activity, decreased food intake, apathy, and lethargy. Many of the mice died, but those that survived were able to gradually resume activities after 4 days. Antibody titer was determined prior to bacterial challenge. Titers of 1:3000 and 1:2000 were measured in the immunized mice using OmpC-EP and OmpC, respectively. No titer was detected in control groups. Significant protective rates were observed in immunized animals, OmpC-EP (62.5%, P < 0.01), and OmpC (56.3%, P < 0.01) (Table 5), indicating that OmpC-EP is effective against bacterial infection; the protective effect of OmpC-EP was slightly higher than that of OmpC.
5. Discussion
There are many methods available for predicting B cell epitopes including BepiPred, ABCpred, and BPAP (16, 17). Tong used the BepiPred 1.0 server to predict 8 B cell epitopes for cockroach (CR) allergies (18). The ABCpred software is mainly based on an artificial neural network algorithm and is highly accurate (13). Using BepiPred and ABCpred, the B cell epitopes were predicted for membrane-associated proteins of Campylobacter jejuni (13) and Gly m Bd 28K of soybean allergens (19). This study used a combination of the BepiPred and ABCpred methods to improve the accuracy of prediction of the B cell epitope for the OmpC protein.
CTL cell epitopes play an integral role in the adaptive immune response (20); and prediction methods include nHLAPred, IEDB, SYFPEITHI, and IMTECH (21-23). Similarly, the nHLAPred method has been used to predict CTL cell epitopes for Leishmania major-related candidate antigens and HIV Gag protein (10, 23), and they showed high immunogenicity. We used the same method to identify a novel critical epitope, which may play a dominant role in activating CD8+T cells (10), and thus increasing the development of immunogenicity.
T cell epitopes have been predicted indirectly by identification of MHC-binding molecules (17). The ProPred method was found to be efficient for Th epitope prediction of bacterial (11, 24) and viral (25) proteins. The predicted Th epitope of the OmpC protein could be combined with MHC-II molecules to enhance the immune response. Epitope joints can be divided into 2 kinds: flexible and rigid (18, 26); frequently-used amino acid connection joints sequences are GGGG, AAY, KK, and GGGGS. We used the GGGG amino acid joint and combined it with the DNAStar software (27) to optimize the different epitopes, and obtained a polypeptide with optimal antigenicity.
Protein antiserum is the basis of immunological function study. The process for preparation of monoclonal antibodies is complex (28), and polyclonal antibodies have the advantages of low cost, shorter time period, and high effect (29). Mouse polyclonal antibodies are commonly used in immunological studies (30). In this study, we obtained a specific antiserum using the method of mouse immunization. The ELISA method is widely used to detect interactions between antigens and antibodies (31, 32). In this study, we observed a direct interaction between OmpC-EP antiserum and E. coli using an optimized ELISA method. When E. coli is combined with antibodies, phagocytes can identify and remove bacteria through their antigen-presenting function (33). Thus, OmpC-EP is expected to show good immunogenicity.
OmpC protein shows good immunogenicity and ability to stimulate the immune system (34, 35). Active immunization with the OmpC protein can enhance the resistance of mice and fish and may have protective functions against different E. coli strains (7, 8). This study designed cell epitope-based polypeptide OmpC-EP to improve the immune function of OmpC protein, and demonstrated that OmpC-EP offers better immune protection than OmpC protein. Thus, OmpC-EP could be used to reduce the E. coli infection and public health concern. However, the development of a recombinant epitope vaccine is still in initial stages, and many problems remain to be solved including the establishment of more effective epitope screening methods and construction of epitope vaccine.
6. Conclusions
In conclusion, a novel cell epitope-based polypeptide, OmpC-EP, for E. coli was designed and confirmed to possess a significant immune protective function. This study is adding a new member to the increasing family of epitope-based vaccine against E. coli infection.
Acknowledgements
References
-
1.
Ravva SV, Sarreal CZ, Mandrell RE. Strain differences in fitness of Escherichia coli O157:H7 to resist protozoan predation and survival in soil. PLoS One. 2014;9(7). e102412. [PubMed ID: 25019377]. https://doi.org/10.1371/journal.pone.0102412.
-
2.
Guo LY, Zhang ZX, Wang X, Zhang PP, Shi W, Yao KH, et al. Clinical and pathogenic analysis of 507 children with bacterial meningitis in Beijing, 2010-2014. Int J Infect Dis. 2016;50:38-43. [PubMed ID: 27452172]. https://doi.org/10.1016/j.ijid.2016.07.010.
-
3.
Keane OM. Genetic diversity, the virulence gene profile and antimicrobial resistance of clinical mastitis-associated Escherichia coli. Res Microbiol. 2016;167(8):678-84. [PubMed ID: 27404459]. https://doi.org/10.1016/j.resmic.2016.06.011.
-
4.
Hussein AH, Ghanem IA, Eid AA, Ali MA, Sherwood JS, Li G, et al. Molecular and phenotypic characterization of Escherichia coli isolated from broiler chicken flocks in Egypt. Avian Dis. 2013;57(3):602-11. [PubMed ID: 24283125]. https://doi.org/10.1637/10503-012513-Reg.1.
-
5.
Hauck CG, Chong PP, Miller MB, Jamieson K, Fine JP, Foster MC, et al. Increasing Rates of Fluoroquinolone Resistance in Escherichia coli Isolated From the Blood and Urine of Patients with Hematologic Malignancies and Stem Cell Transplant Recipients. Pathog Immun. 2016;1(2):234-42. [PubMed ID: 28004038]. https://doi.org/10.20411/pai.v1i2.115.
-
6.
Letuta UG, Vekker AS, Kornilova TA, Gryaznov AA, Cheplakov IA. Magnetic isotope effect of magnesium (25)Mg on E. coli resistance to antibiotics. Dokl Biochem Biophys. 2016;469(1):281-3. [PubMed ID: 27599512]. https://doi.org/10.1134/S1607672916040128.
-
7.
Liu C, Chen Z, Tan C, Liu W, Xu Z, Zhou R, et al. Immunogenic characterization of outer membrane porins OmpC and OmpF of porcine extraintestinal pathogenic Escherichia coli. FEMS Microbiol Lett. 2012;337(2):104-11. [PubMed ID: 23003111]. https://doi.org/10.1111/1574-6968.12013.
-
8.
Perez-Toledo M, Valero-Pacheco N, Pastelin-Palacios R, Gil-Cruz C, Perez-Shibayama C, Moreno-Eutimio MA, et al. Salmonella Typhi Porins OmpC and OmpF Are Potent Adjuvants for T-Dependent and T-Independent Antigens. Front Immunol. 2017;8:230. [PubMed ID: 28337196]. https://doi.org/10.3389/fimmu.2017.00230.
-
9.
Ikram A, Anjum S, Tahir M. In Silico Identification and Conservation Analysis of B-cell and T-Cell Epitopes of Hepatitis C Virus 3a Genotype Enveloped Glycoprotein 2 From Pakistan: A Step Towards Heterologous Vaccine Design. Hepat Mon. 2014;14(6). e9832. [PubMed ID: 24976845]. https://doi.org/10.5812/hepatmon.9832.
-
10.
Abidi SH, Shahid A, Lakhani LS, Khanani MR, Ojwang P, Okinda N, et al. Population-specific evolution of HIV Gag epitopes in genetically diverged patients. Infect Genet Evol. 2013;16:78-86. [PubMed ID: 23403357]. https://doi.org/10.1016/j.meegid.2013.02.003.
-
11.
Saraav I, Pandey K, Sharma M, Singh S, Dutta P, Bhardwaj A, et al. Predicting promiscuous antigenic T cell epitopes of Mycobacterium tuberculosis mymA operon proteins binding to MHC Class I and Class II molecules. Infect Genet Evol. 2016;44:182-9. [PubMed ID: 27389362]. https://doi.org/10.1016/j.meegid.2016.07.004.
-
12.
Diken M, Vormehr M, Grunwitz C, Kreiter S, Tureci O, Sahin U. Discovery and Subtyping of Neo-Epitope Specific T-Cell Responses for Cancer Immunotherapy: Addressing the Mutanome. Methods Mol Biol. 2017;1499:223-36. [PubMed ID: 27987153]. https://doi.org/10.1007/978-1-4939-6481-9_14.
-
13.
Yasmin T, Akter S, Debnath M, Ebihara A, Nakagawa T, Nabi AH. In silico proposition to predict cluster of B- and T-cell epitopes for the usefulness of vaccine design from invasive, virulent and membrane associated proteins of C. jejuni. In Silico Pharmacol. 2016;4(1):5. [PubMed ID: 27376537]. https://doi.org/10.1186/s40203-016-0020-y.
-
14.
Liu X, She XT, Zhu QF, Li H, Peng XX. Heterogeneous interactome between Litopenaeus vannamei plasma proteins and Vibrio parahaemolyticus outer membrane proteins. Fish Shellfish Immunol. 2013;34(1):192-8. [PubMed ID: 23099052]. https://doi.org/10.1016/j.fsi.2012.10.023.
-
15.
Liu Y, Zhang H, Liu Y, Li H, Peng X. Determination of the heterogeneous interactome between Edwardsiella tarda and fish gills. J Proteomics. 2012;75(4):1119-28. [PubMed ID: 22079874]. https://doi.org/10.1016/j.jprot.2011.10.022.
-
16.
Jafarpour S, Ayat H, Ahadi AM. Design and Antigenic Epitopes Prediction of a New Trial Recombinant Multiepitopic Rotaviral Vaccine: In Silico Analyses. Viral Immunol. 2015;28(6):325-30. [PubMed ID: 25965449]. https://doi.org/10.1089/vim.2014.0152.
-
17.
Yang H, Chen H, Jin M, Xie H, He S, Wei JF. Molecular cloning, expression, IgE binding activities and in silico epitope prediction of Per a 9 allergens of the American cockroach. Int J Mol Med. 2016;38(6):1795-805. [PubMed ID: 27840974]. https://doi.org/10.3892/ijmm.2016.2793.
-
18.
Tong X, Guo M, Jin M, Chen H, Li Y, Wei JF. In silico epitope prediction, expression and functional analysis of Per a 10 allergen from the American cockroach. Int J Mol Med. 2016;38(6):1806-14. [PubMed ID: 27840898]. https://doi.org/10.3892/ijmm.2016.2790.
-
19.
Chabner BA, Shoemaker D. Drug development for cancer: implications for chemical modifiers. Int J Radiat Oncol Biol Phys. 1989;16(4):907-9. [PubMed ID: 2703396].
-
20.
Cui T, Yi X, Guo S, Zhou F, Liu L, Li C, et al. Identification of Novel HLA-A*0201-Restricted CTL Epitopes in Chinese Vitiligo Patients. Sci Rep. 2016;6:36360. [PubMed ID: 27821860]. https://doi.org/10.1038/srep36360.
-
21.
de la Poza F, Marin-Lopez A, Castillo-Olivares J, Calvo-Pinilla E, Ortego J. Identification of CD8 T cell epitopes in VP2 and NS1 proteins of African horse sickness virus in IFNAR(-/-) mice. Virus Res. 2015;210:149-53. [PubMed ID: 26272673]. https://doi.org/10.1016/j.virusres.2015.08.005.
-
22.
Ma RX, Cheng LF, Ying QK, Liu RR, Ma TJ, Zhang XX, et al. Screening and Identification of an H-2Kb-Restricted CTL Epitope within the Glycoprotein of Hantaan Virus. Front Cell Infect Microbiol. 2016;6:151. [PubMed ID: 27933274]. https://doi.org/10.3389/fcimb.2016.00151.
-
23.
Seyed N, Zahedifard F, Safaiyan S, Gholami E, Doustdari F, Azadmanesh K, et al. In silico analysis of six known Leishmania major antigens and in vitro evaluation of specific epitopes eliciting HLA-A2 restricted CD8 T cell response. PLoS Negl Trop Dis. 2011;5(9). e1295. [PubMed ID: 21909442]. https://doi.org/10.1371/journal.pntd.0001295.
-
24.
Rauta PR, Ashe S, Nayak D, Nayak B. In silico identification of outer membrane protein (Omp) and subunit vaccine design against pathogenic Vibrio cholerae. Comput Biol Chem. 2016;65:61-8. [PubMed ID: 27769003]. https://doi.org/10.1016/j.compbiolchem.2016.10.004.
-
25.
Dar H, Zaheer T, Rehman MT, Ali A, Javed A, Khan GA, et al. Prediction of promiscuous T-cell epitopes in the Zika virus polyprotein: An in silico approach. Asian Pac J Trop Med. 2016;9(9):844-50. [PubMed ID: 27633296]. https://doi.org/10.1016/j.apjtm.2016.07.004.
-
26.
Leroux LP, Dasanayake D, Rommereim LM, Fox BA, Bzik DJ, Jardim A, et al. Secreted Toxoplasma gondii molecules interfere with expression of MHC-II in interferon gamma-activated macrophages. Int J Parasitol. 2015;45(5):319-32. [PubMed ID: 25720921]. https://doi.org/10.1016/j.ijpara.2015.01.003.
-
27.
He J, Zhang J, He Y, Huang F, Li J, Chen Q, et al. Construction of recombinant Mip-FlaA dominant epitope vaccine against Legionella pneumophila and evaluation of the immunogenicity and protective immunity. Immunol Res. 2016;64(1):272-9. [PubMed ID: 26607265]. https://doi.org/10.1007/s12026-015-8746-x.
-
28.
Miller BJ, Buckley PF. Monoclonal antibody immunotherapy in psychiatric disorders. Lancet Psychiatry. 2017;4(1):13-5. [PubMed ID: 28012468]. https://doi.org/10.1016/S2215-0366(16)30366-2.
-
29.
Doria-Rose NA, Altae-Tran HR, Roark RS, Schmidt SD, Sutton MS, Louder MK, et al. Mapping Polyclonal HIV-1 Antibody Responses via Next-Generation Neutralization Fingerprinting. PLoS Pathog. 2017;13(1). e1006148. [PubMed ID: 28052137]. https://doi.org/10.1371/journal.ppat.1006148.
-
30.
Zheng Q, Wang T, Jiang S, Han R, Jin N, Zhu J, et al. Production of Polyclonal Antibody to the HPV58 E7 Protein and Its Detection in Cervical Cancer. PLoS One. 2016;11(12). e0169138. [PubMed ID: 28033368]. https://doi.org/10.1371/journal.pone.0169138.
-
31.
Petruccioli E, Vanini V, Chiacchio T, Cirillo DM, Palmieri F, Ippolito G, et al. Modulation of interferon-gamma response to QuantiFERON-TB-plus detected by enzyme-linked immunosorbent assay in patients with active and latent tuberculosis infection. Int J Mycobacteriol. 2016;5 Suppl 1:S143-4. [PubMed ID: 28043514]. https://doi.org/10.1016/j.ijmyco.2016.09.029.
-
32.
Teymouri H, Mosavari N, Poor Taghi H. Detection of Mycobacterium avium subsp. Paratuberculosis in Cattle by using Indirect Absorbed ELISA (enzyme-linked immunosorbent assay) system and culture in Alborz Province, Iran. Int J Mycobacteriol. 2016;5 Suppl 1:S220-1. [PubMed ID: 28043567]. https://doi.org/10.1016/j.ijmyco.2016.10.006.
-
33.
Novakowski KE, Loukov D, Chawla V, Bowdish DM. Bacterial Binding, Phagocytosis, and Killing: Measurements Using Colony Forming Units. Methods Mol Biol. 2017;1519:297-309. [PubMed ID: 27815888]. https://doi.org/10.1007/978-1-4939-6581-6_20.
-
34.
Thulasingam M, Damodharan S, Madhana Vigneshwari G, P. J. Pandaranayaka E, Elizabeth Hanna L, Usha R, et al. Characterization of Salmonella typhi OmpC and OmpF porins engineered with HIV-gp41 epitope on the surface loops. Proteins. 2017;85(4):657-64. [PubMed ID: 28076882]. https://doi.org/10.1002/prot.25246.
-
35.
Xiao XH, Miao HM, Xu YG, Zhang JY, Chai LH, Xu JJ. Analysis of skin and secretions of Dybowski's frogs (Rana dybowskii) exposed to Staphylococcus aureus or Escherichia coli identifies immune response proteins. Vet J. 2014;200(1):127-32. [PubMed ID: 24613415]. https://doi.org/10.1016/j.tvjl.2014.01.011.