Abstract
Background:
Methicillin-resistant Staphylococcus aureus (MRSA) continues to be a major cause of nosocomial infections. Methicillin resistance in S. aureus is caused by the acquisition of the mecA gene, located on a mobile genetic element called the staphylococcal cassette chromosome (SCC).Objectives:
The aim of this study was to evaluate the presence of the predominant SCCmec type present among clinical isolates.Materials and Methods:
This cross-sectional study was performed on a total of 146 MRSA isolates obtained from clinical specimens between 2012 and 2013 from two major hospitals in Shiraz, Southwest of Iran. Antibiotic susceptibility profiles were determined by the disc diffusion method according to the guidelines of The Clinical and Laboratory Standards Institute. Bacterial DNA was extracted using the small-scale phenol-chloroform extraction method and was employed as polymerase chain reaction (PCR) templates for the assigned current SCCmec types.Results:
The assigned SCCmec types by PCR revealed the SCCmec type I as the predominant type with 86 (58.9%) samples, followed by the SCCmec type II with 29 (19.9%), type III with 16 (11.0%), and type IV with 12 (8.2%) samples, respectively. The SCCmec type I MRSA isolates were significantly recovered from blood (80%) and sputum (67.2%). The results of antibacterial susceptibility tests for the MRSA isolates showed that all of those carrying the SCCmec type I and II had significantly greater resistance rates to Gentamicin and Rifampin than the isolates containing the SCCmec type III. Also, a significant difference was detected for susceptibility to Co-trimoxazole between the SCCmec type I and II MRSA isolates and the SCCmec type III, which was more resistant.Conclusions:
The frequency of the isolates containing type I in the current study can indicate an emergence of this SCCmec type in the studied medical centers.Keywords
Methicillin-Resistant Staphylococcus aureus (MRSA) Mobile Genetic Elements Antimicrobial Susceptibility
1. Background
Methicillin-resistant Staphylococcus aureus (MRSA) continues to be a major cause of nosocomial infections as it adapts rapidly to selective pressures imparted by the human host (1). Mobile genetic elements play a central role in this adaptation process by encoding virulence factors and molecules that confer resistance to antibiotics, including the gene that confers resistance to beta-lactam antibiotics in MRSA (1). Methicillin resistance in S. aureus is caused by the expression of PBP2a encoded by the mecA gene, which is located on a genetic element called the staphylococcal cassette chromosome (SCC) (2).The SCCmec is a group of mobile DNA elements integrated into the chromosome of MRSA. To date, 11 types of the SCCmec have been reported (2). The distribution of the 3 types of SCCmec (i.e. types I, II, and III) in the world has been shown, and they were originally described in hospital-acquired MRSA isolates (3-5). Also, types IV and V have been reported in a community-acquired MRSA isolate (5, 6). The rest of them were reported outspread in some areas (2). MRSA isolated from hospital-acquired MRSA is usually resistant to multiple antibiotics, and it typically contains larger SCCmec.
Nosocomial infections are an important health problem worldwide due to multidrug-resistant MRSA (7, 8). Our understanding from MRSA typing and resistance genes which it expresses and the pattern of its antimicrobial resistance can provide useful information toward optimizing infection control and clinical therapy in the future. In light of previous data showing that all MRSA isolates contain specific types of the SCCmec, sequence-based methods can be used to compare the results obtained at different places and times and find their relationship with the origin of the infections (2). The emergence of MRSA from Iran with a higher prevalence compared to the reports from neighboring countries in the Middle East calls for a great and urgent effort to understand MRSA resistance mechanisms and its current antimicrobial profile for the prevention and the selection of appropriate treatment (9).
2. Objectives
The aim of this study was to evaluate the presence of predominant SCCmec type/s present among clinical isolates from our regional hospitals in a large city in the southwest of Iran, Shiraz.
3. Materials and Methods
3.1. Bacterial Strains
This cross-sectional study was conducted between October 2012 and March 2013 from two hospitals, Nemazee and Faghihi, in Shiraz, a major city in the southwest of Iran. Nemazee and Faghihi hospitals are two major tertiary care hospitals with 1000 beds, affiliated to Shiraz University of Medical Sciences, Shiraz, Iran. A total of 345 S. aureus isolates were obtained from the clinical specimens of the two mentioned hospitals. Specimens were collected from different wards in the hospitals. Outpatient specimens and duplicate isolates were not included. The isolates were identified as S. aureus using standard procedures (colony morphology, Gram stain, catalase activity, growth on Mannitol salt agar, DNase test, and tube coagulase) and stored at -70°C for long-term preservation.
The MRSA isolates were screened based on resistance to Cefoxitin (30 μg) discs (MAST, UK) by the disc diffusion method according to the guidelines of The Clinical and Laboratory Standards Institute (CLSI) (10). The isolates resistant to Cefoxitin were further tested for the presence of the mecA gene. Totally, 146 confirmed MRSA isolates were obtained from the sputum (58), blood (20), wound (18), urine (14), pus (8), endotracheal tube (6), and 22 other specimens.
3.2. Antimicrobial Susceptibility Testing
Susceptibility profiles were determined to Ampicillin, Gentamicin, Erythromycin, Tetracycline, Ciprofloxacin, Clindamycin, Teicoplanin, Co-Trimoxazole (SXT), Chloramphenicol, Rifampicin, Linezolid, Synercid®, and Vancomycin antibiotic discs (MAST, UK) by the disc diffusion method according to the guidelines of the CLSI on Muller-Hinton agar (Oxoid, UK). S. aureus ATCC 25923, which is a Methicillin-sensitive S. aureus, was used in this study as the control strain in antibacterial susceptibility testing. The interpretation of the diameters of the zones of inhibition was done according to the guidelines of the CLSI (10). Inducible Clindamycin resistance was also detected using the D-test method described by the guidelines of the CLSI (10).
3.3. DNA Extraction and Molecular Typing
Bacterial whole DNA was extracted from the MRSA isolates using the small-scale phenol-chloroform extraction method and was utilized as polymerase chain reaction (PCR) templates (11). The SCCmec typing was optimized to use PCR experiments via a multiplex PCR protocol described for the SCCmec types I-V by Zhang et al. (12), with the exception of the SCCmec type IV, which was used as a specific primer designed by Oliveira and de Lencastre (13). Also, the mecA gene detection was amplified using a primer described by Zhang et al. (12). All the primers used in the current study were manufactured by CinnaGen, Iran.
The PCR amplifications were performed in a DNA Thermal Cycler 5530 (Eppendorf Master, Germany). The PCR products were resolved by electrophoresis in 3% agarose gels at 70V and band was observed by UV irradiation (Figure 1). The following MRSA reference strains were used as controls: COL; N315; 85/2082; and JCSC/4469 for the SCCmec types I, II, III, and IV, respectively. All the reference strains were kindly provided by the Professor Alborzi Clinical Microbiology Research Center, Shiraz, Iran. The DNA sequencing was performed (BIONEER, South Korea) to confirm the presence of the SCCmec I genes in a subset of 3 positive isolates for the SCCmec type I in PCR.Statistical analysis was performed using the SPSS software (version 19.0). The chi-square test or the Fisher’s exact test was conducted to analyze the results. A P value < 0.05 was regarded as statistically significant.
A Representative Gel Image of SCCmec Typing by Polymerase Chain Reaction
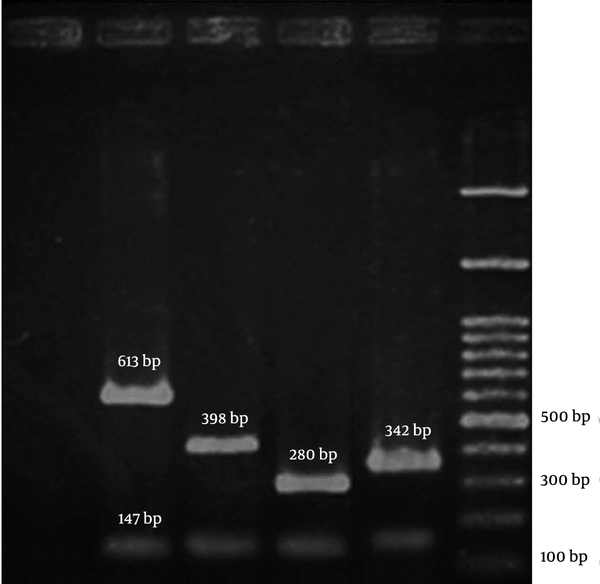
4. Results
Of the 345 S. aureus isolates obtained from Nemazee and Faghihi hospitals between 2012 and 2013, 146 (42.3%) were Methicillin resistant according to the disc diffusion and the mecA analysis.
For all the 146 MRSA isolates, the SCCmec types were assigned by PCR. It was revealed that the SCCmec type I was the predominant type with 86 (58.9%) samples, followed by the SCCmec type II with 29 (19.9%), type III with 16 (11.0%), and type IV with 12 (8.2%) samples, respectively. Three (2.1%) MRSA isolates remained untypeable. The SCCmec type I MRSA isolates were significantly (P < 0.001) recovered as the predominant type from 16 (80.0%) blood and 39 (67.2%) sputum samples, while the SCCmec type II isolates were mostly (13 [44.8%]) isolated from the sputum samples (P < 0.001). Also, most of the MRSA isolates recovered from skin belonged to the SCCmec types II and III (P < 0.001) (Table 1).
Frequency of Different SCCmec Types of Methicillin-Resistant Staphylococcus aureus Isolated From Clinical Specimens a
| SCCmec Types, Source | I, n = 86 | II, n = 29 | III, n = 16 | IV, n = 12 | Untypeable, n = 3 | Total, n = 146 |
|---|---|---|---|---|---|---|
| Sputum b,c | 39 (45.3) | 13 (44.8) | 3 (18.7) | 2 (16.7) | 1 (33.3) | 58 (39.7) |
| Blood b | 16 (18.6) | 1 (3.4) | 1 (6.3) | 2 (16.7) | 0 (0) | 20 (13.7) |
| Wound | 10 (11.6) | 5 (17.2) | 1 (6.3) | 2 (16.7) | 0 (0) | 18 (12.3) |
| Urine | 5 (5.8) | 2 (6.9) | 3 (18.7) | 3 (25.0) | 1 (33.3) | 14 (9.6) |
| Skin d | 1 (1.2) | 2 (6.9) | 4 (25.0) | 1 (8.3) | 0 (0) | 8 (5.5) |
| Endotracheal tube | 2 (2.3) | 0 (0) | 2 (12.5) | 1 (8.3) | 1 (33.3) | 6 (4.1) |
| Other e | 13 (15.1) | 6 (20.7) | 2 (12.5) | 1 (8.3) | 0 (0) | 22 (15.1) |
There were no significant differences in the detected SCCmec type frequencies between the two hospitals, except for the SCCmec type II MRSA isolates, which had a higher prevalence rate in Faghihi Hospital (P < 0.05). The results of antibacterial susceptibility tests for the MRSA isolates showed that all the isolates were resistant to Ampicillin. Moreover, all of them were sensitive to Linezolid, Teicoplanin, Synercid®, and Vancomycin (Table 2).
| Resistance, Antibiotics | I, n = 86 | II, n = 29 | III, n = 16 | IV b, n = 12 | Untypeable, n = 3 | Total, n = 146 |
|---|---|---|---|---|---|---|
| Ciprofloxacin | 85 (98.8) | 29 (100) | 16 (100) | 0 (0) | 0 (0) | 130 (89.0) |
| Gentamycin c | 84 (97.7) | 28 (96.6) | 12 (75.0) | 0 (0) | 0 (0) | 124 (84.9) |
| Clindamycin | 83 (96.5) | 25 (86.2) | 16 (100) | 3 (25.0) | 1 (33.3) | 128 (87.7) |
| Erythromycin | 83 (96.5) | 25 (86.2) | 16 (100) | 3 (25.0) | 1 (33.3) | 128 (87.7) |
| Tetracycline | 79 (91.9) | 28 (96.6) | 16 (100) | 6 (50.0) | 2 (66.7) | 131 (89.7) |
| Rifampin c | 70 (81.4) | 21 (72.4) | 6 (37.5) | 0 (0) | 0 (0) | 97 (66.4) |
| Co-trimoxazole d | 16 (18.6) | 7 (24.1) | 11 (68.7) | 0 (0) | 0 (0) | 34 (23.3) |
| Chloramphenicol | 2 (2.3) | 1 (3.4) | 0 (0) | 0 (0) | 1 (33.3) | 4 (2.7) |
Among the clinical specimens, the isolates recovered from skin showed a significantly higher resistance rate than the urine samples (P < 0.05). The MRSA isolates that carried the SCCmec types I and II revealed a significantly greater resistance to Gentamicin and Rifampin than the isolates containing the SCCmec type III (P < 0.05). Also, a significant difference was seen for susceptibility to SXT between the SCCmec types I and II MRSA isolates and the SCCmec type III, which was more resistant (P < 0.05). Overall, the MRSA isolates containing the SCCmec type IV were more sensitive than the other types, especially to antibiotics including Ciprofloxacin, Gentamicin, Rifampin, SXT, and Chloramphenicol. Also, multidrug-resistance was significantly lower in the SCCmec type IV than in the SCCmec types I, II, and III, which were mostly resistant to more than 5 antibiotics (P < 0.001) (Figure 2).
Comparison of the Prevalence of Multidrug-Resistant Isolates between the Different SCCmec Types
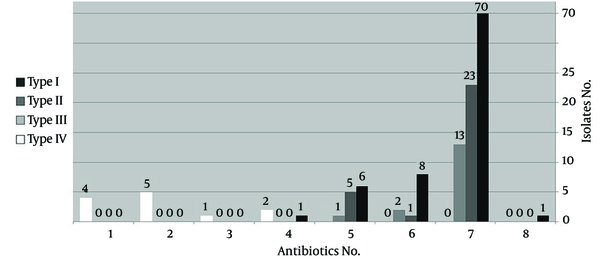
The MRSA isolates belonging to the SCCmec type I (66.3%) and type II (65.5%) were greatly recovered from patients in the Intensive Care Unit (ICU), while the SCCmec type III was mostly recovered from patients hospitalized on other wards (P < 0.001). The D-test results revealed that 14 MRSA isolates had inducible resistance to Clindamycin. There was a significant difference regarding the number of the inducible isolates recovered from the two hospitals (13 (92.9%) samples from Faghihi Hospital vs. 1 (7.1%) sample from Nemazee Hospital, P < 0.001). Also, a significant correlation was observed between susceptibility to Rifampin and inducible Clindamycin resistance because the 13 isolates with inducible resistance remained sensitive to Rifampin (P < 0.001). DNA sequencing confirmed the presence of the SCCmec type I gene with checking through the SCCmec database at www.staphylococcus.net/ and using The National Center for Biotechnology Information's (NCBI) blast tools, which also revealed that all the isolates were closely related to the COL MRSA strain.
5. Discussion
In this study, the prevalence of MRSA among the S. aureus isolates was found to be 42.3% (146), slightly higher than that in similar reports from the north of Iran, Tehran with 36% (3). However, the prevalence of MRSA varies widely among different regions and countries; this fact reflects the different infection control policies and other factors involved. Most of the specimens in this study were isolated from sputum (58 = 39.7%) and blood (20 = 13.7%). It is in consistence with the previous studies reporting that MRSA isolates are responsible for the majority of respiratory and blood infections (septicemia, endocarditic, etc.) in hospitalized patients (8, 14, 15). Also, a higher frequency of respiratory infections in those studies can indicate that most of the MRSA nosocomial transmissions occurred by the respiratory tract (7, 16).
In this study, the antibiotic susceptibility pattern of the isolates to 13 commonly used antibiotics for MRSA infections was also evaluated. The sensitivity of the entire MRSA isolates to Vancomycin, Linezolid, Teicoplanin, and Synercid® in this study was similar to that in most reports, so these agents can be used as therapeutic choices in our medical centers. Of course, the cost-benefitness and the different pharmacological properties of these agents should be taken into consideration when prescribing antibiotics (8, 17). Chloramphenicol and SXT with sensitivity rates of 97.3% and 76.7%, respectively among our studied isolates, could be considered as the subsequent treatment options. Also, the skin MRSA isolates were more resistant than those of the urine isolates significantly. This may be a reflection of unique bacterial exposure with environmental or specific physiological conditions of these areas. The role of environmental factors such as nurses’ hands and clothes in the transmission of resistant isolates on the patient’s skin in various studies has been shown previously (18, 19).
In this study, the frequency of induced Clindamycin resistance was detected to be 9.6% among the MRSA isolates. In previous reports in Iran, inducible Clindamycin resistance for MRSA isolates ranged between 2.3% and 33.3% (20-22). Also, this frequency has been reported in various rates depending on the geographical distribution from some Asian countries (23, 24). From a total of 146 MRSA isolates tested in this study, the predominant SCCmec types were the SCCmec type I with a rate of 58.9% (n = 86) and the SCCmec type II with a rate of 19.9% (29). These data were similar to the results from a study by Moghadami et al. (4), who reported type I and II isolation rates of 56.9% and 22%, respectively, as the prevalent SCCmec types from 109 MRSA isolates collected from Tehran and Shiraz, Iran. Also, in a hospital survey in the neighboring country, Pakistan, in 2010, the SCCmec types I and II were the predominant isolates with 60% and 40% rates, respectively (25). Some other studies in Iran have also reported the relative prevalence of the SCCmec types I and II (26, 27). Fatholahzadeh et al. (3) and Japoni et al. (8) in two related studies in Iran reported the SCCmec type III as the most predominant type: rates of 98% in the former and 74.3% in the latter. According to previous reports, most of the hospital-acquired MRSA isolates were assigned as the SCCmec types I to III, which is consistent with our results (3, 4, 28).
The majority of the type I isolates in the present study were from the blood and sputum specimens. It has been previously observed that some plasmin-sensitive surface proteins are located in the specific regions of the SCCmec type I elements and are associated with a more efficient distribution of bacteria throughout the body, especially the circulatory system. In some studies, the role of type I MRSA isolates in blood-related diseases has been shown (29, 30). Moreover, in the current study, most of the isolates containing the SCCmec type II were isolated from the sputum specimens (44.8%), which is in agreement with the previous investigations suggesting the high frequency of this SCCmec-type isolates from respiratory infections (7). Furthermore, as was mentioned, the prevalence rates of type I (66.3%) and II (65.5%) MRSA isolates were significantly higher (P < 0.001) in the ICU. It may reflect the fact that some patients such as critically ill patients in the ICU have a greater chance of colonization and infection by these specific SCCmec types (31).
It has been already shown that the SCCmec types I to III are frequently isolated from hospital-acquired MRSA and often apart from the mecA gene containing additional genes for resistance to several beta-lactam antibiotics (2). Several reasons to gain additional resistance genes by these isolates have been reported in medical centers (8, 32), which could explain the high antibiotic resistance of the SCCmec type I-III isolates in our study. Also, the significant antibiotic susceptibility pattern of certain SCCmec type isolates in our results can prompt us to consider these antibiotics as the treatment option for the infection that caused these isolates. In the present study, the majority (about 70 - 80%) of the SCCmec type I and II isolates showed susceptibility to SXT, which may be considered to be connected clonally with these isolates (33). Furthermore, the high prevalence of the SCCmec type I isolates observed in the current study with a significantly higher multidrug-resistance (to 7 and 6 antimicrobial agents) indicates that they may adapt and survive in our regional hospitals over time.
Most of the type IV isolates in our study were susceptible to the majority of the tested antibiotics (9 - 10 agents). However, as is indicated in some studies, a few isolates containing type IV cassette were multidrug-resistant in our study as well (7, 8). This suggests that since type IV isolates are often exposed to antibiotics, they could gain antibiotic resistance genes other than the mecA gene to survive in medical centers (8, 34). The frequency of the SCCmec type I isolates in the present study can indicate the emergence of this type in the studied medical centers. As was previously mentioned in our results, due to the significant clinical source and the antimicrobial susceptibility pattern of different SCCmec type isolates, optimizing institutional infection control policies for preventing MRSA transmission among hospitalized patients should be considered. Our findings highlight the importance of the continual monitoring of molecular changes with regard to the related susceptibility patterns of isolates to commonly clinically used antibiotics in our region.
Acknowledgements
References
-
1.
Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010;67(18):3057-71. [PubMed ID: 20668911]. https://doi.org/10.1007/s00018-010-0389-4.
-
2.
Turlej A, Hryniewicz W, Empel J. Staphylococcal cassette chromosome mec (Sccmec) classification and typing methods: an overview. Pol J Microbiol. 2011;60(2):95-103. [PubMed ID: 21905625].
-
3.
Fatholahzadeh B, Emaneini M, Gilbert G, Udo E, Aligholi M, Modarressi MH, et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist. 2008;14(3):217-20. [PubMed ID: 18694326]. https://doi.org/10.1089/mdr.2008.0822.
-
4.
Moghadami M, Japoni A, Karimi A, Mardani M. Comparison of community and healthcare-associated MRSA in Iran. Arch Clin Infect Dis. 2010;5(4):206-12.
-
5.
Naimi TS, LeDell KH, Como-Sabetti K, Borchardt SM, Boxrud DJ, Etienne J, et al. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003;290(22):2976-84. [PubMed ID: 14665659]. https://doi.org/10.1001/jama.290.22.2976.
-
6.
Maree CL, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG. Community-associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerg Infect Dis. 2007;13(2):236-42. [PubMed ID: 17479885]. https://doi.org/10.3201/eid1302.060781.
-
7.
Kilic A, Li H, Stratton CW, Tang YW. Antimicrobial susceptibility patterns and staphylococcal cassette chromosome mec types of, as well as Panton-Valentine leukocidin occurrence among, methicillin-resistant Staphylococcus aureus isolates from children and adults in middle Tennessee. J Clin Microbiol. 2006;44(12):4436-40. [PubMed ID: 17065272]. https://doi.org/10.1128/JCM.01546-06.
-
8.
Japoni A, Jamalidoust M, Farshad S, Ziyaeyan M, Alborzi A, Japoni S, et al. Characterization of SCCmec types and antibacterial susceptibility patterns of methicillin-resistant Staphylococcus aureus in Southern Iran. Jpn J Infect Dis. 2011;64(1):28-33. [PubMed ID: 21266752].
-
9.
Askari E, Soleymani F, Arianpoor A, Tabatabai SM, Amini A, Naderinasab M. Epidemiology of mecA-Methicillin Resistant Staphylococcus aureus (MRSA) in Iran: A Systematic Review and Meta-analysis. Iran J Basic Med Sci. 2012;15(5):1010-9. [PubMed ID: 23493646].
-
10.
Performance standards for antimicrobial susceptibility testing; 21th Informational Supplement. Wayne,: CLSI; 2011.
-
11.
Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc. 2006;2006(1). [PubMed ID: 22485786]. https://doi.org/10.1101/pdb.prot4455.
-
12.
Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026-33. [PubMed ID: 16207957]. https://doi.org/10.1128/JCM.43.10.5026-5033.2005.
-
13.
Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155-61. [PubMed ID: 12069968].
-
14.
Bereket W, Hemalatha K, Getenet B, Wondwossen T, Solomon A, Zeynudin A, et al. Update on bacterial nosocomial infections. Eur Rev Med Pharmacol Sci. 2012;16(8):1039-44. [PubMed ID: 22913154].
-
15.
Tacconelli E. Antimicrobial use: risk driver of multidrug resistant microorganisms in healthcare settings. Curr Opin Infect Dis. 2009;22(4):352-8. [PubMed ID: 19461514]. https://doi.org/10.1097/QCO.0b013e32832d52e0.
-
16.
Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J Infect Dis. 2006;193(2):172-9. [PubMed ID: 16362880]. https://doi.org/10.1086/499632.
-
17.
Gould FK, Brindle R, Chadwick PR, Fraise AP, Hill S, Nathwani D, et al. Guidelines (2008) for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the United Kingdom. J Antimicrob Chemother. 2009;63(5):849-61. [PubMed ID: 19282331]. https://doi.org/10.1093/jac/dkp065.
-
18.
Morgan DJ, Rogawski E, Thom KA, Johnson JK, Perencevich EN, Shardell M, et al. Transfer of multidrug-resistant bacteria to healthcare workers' gloves and gowns after patient contact increases with environmental contamination. Crit Care Med. 2012;40(4):1045-51. [PubMed ID: 22202707]. https://doi.org/10.1097/CCM.0b013e31823bc7c8.
-
19.
Sahoo K, Tamhankar AJ, Johansson E, Lundborg C. Antibiotic use, resistance development and environmental factors: a qualitative study among healthcare professionals in Orissa, India. BMC Public Health. 2010;10(1):629. https://doi.org/10.1186/1471-2458-10-629.
-
20.
Seifi N, Kahani N, Askari E, Mahdipour S, Naderi NM. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iran J Microbiol. 2012;4(2):82-6. [PubMed ID: 22973474].
-
21.
Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med Sci Monit. 2011;17(2):BR48-53. [PubMed ID: 21278685].
-
22.
Memariani M, Pourmand MR, Shirazi MH, Dallal MMS, Abdossamadi Z, Mardani N. The importance of inducible clindamycin resistance in enterotoxin positive S. aureus isolated from clinical samples. Tehran Uni Med J. 2009;67(4):250-6.
-
23.
Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol. 2004;42(6):2777-9. [PubMed ID: 15184468]. https://doi.org/10.1128/JCM.42.6.2777-2779.2004.
-
24.
Velvizhi G, Sucilathangam G, Palaniappan N. Prevalence and phenotypic detection of erythromycin-induced resistance to clindamycin in MRSA isolates. J Clin Diagn Res. 2011;5(6):1195-8.
-
25.
Romeeza T, Naeem R, Nadeem A. Multiplex Polymerase Chain Reaction Based Typing of Staphylococcal Chromosomal Cassette mec of Methicillin Resistant Staphylococcus aureus from Selected Hospitals in Lahore. I Med J Malaysia. 2010;9(1).
-
26.
Zeinali E, Moniri R, Musavi GH. Antibiotic resistance and molecular subtypes of clinical methicillin-resistant Staphylococcus aureus in a teaching hospital. Indian J Med Microbiol. 2011;29(3):318-9. [PubMed ID: 21860123]. https://doi.org/10.4103/0255-0857.83926.
-
27.
Sadeghi J, Mansouri S. Molecular characterization and antibiotic resistance of clinical isolates of methicillin-resistant Staphylococcus aureus obtained from Southeast of Iran (Kerman). APMIS. 2014;122(5):405-11. [PubMed ID: 24033803]. https://doi.org/10.1111/apm.12158.
-
28.
Kim JS, Song W, Kim HS, Cho HC, Lee KM, Choi MS, et al. Association between the methicillin resistance of clinical isolates of Staphylococcus aureus, their staphylococcal cassette chromosome mec (SCCmec) subtype classification, and their toxin gene profiles. Diagn Microbiol Infect Dis. 2006;56(3):289-95. [PubMed ID: 16854552]. https://doi.org/10.1016/j.diagmicrobio.2006.05.003.
-
29.
Werbick C, Becker K, Mellmann A, Juuti KM, von Eiff C, Peters G, et al. Staphylococcal chromosomal cassette mec type I, spa type, and expression of Pls are determinants of reduced cellular invasiveness of methicillin-resistant Staphylococcus aureus isolates. J Infect Dis. 2007;195(11):1678-85. [PubMed ID: 17471438]. https://doi.org/10.1086/517517.
-
30.
Josefsson E, Juuti K, Bokarewa M, Kuusela P. The surface protein Pls of methicillin-resistant Staphylococcus aureus is a virulence factor in septic arthritis. Infect Immun. 2005;73(5):2812-7. [PubMed ID: 15845485]. https://doi.org/10.1128/IAI.73.5.2812-2817.2005.
-
31.
Fluit AC, Wielders CL, Verhoef J, Schmitz FJ. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. J Clin Microbiol. 2001;39(10):3727-32. [PubMed ID: 11574603]. https://doi.org/10.1128/JCM.39.10.3727-3732.2001.
-
32.
Aiello AE, Larson E. Antibacterial cleaning and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect Dis. 2003;3(8):501-6. https://doi.org/10.1016/s1473-3099(03)00723-0.
-
33.
Oliveira DC, Tomasz A, De-Lencastre H. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002;2(3):180-9.
-
34.
Takata T, Shirakawa T, Ito J, Okamoto A, Massi MN, Kinoshita S, et al. SCCmec typing and detection of VISA-related genes in methicillin-resistant Staphylococcus aureus clinical strains from Kobe University Hospital, Japan. Southeast Asian J Trop Med Public Health. 2006;37(6):1149-55. [PubMed ID: 17333768].