Abstract
Background:
PCR is a molecular technique for herpes simplex virus (HSV) detection that can cause life-threatening infections such as encephalitis and keratitis. However, the main issues, false-negative results causing by PCR inhibitors, of this technique that reduce PCR efficiency. To overcome this problem, a competitive internal amplification control (IAC) was constructed for conventional PCR using the PCR-cloning technique.Objectives:
The purpose of this study is the design of competitive IAC for PCR diagnosis of HSV, which in fact is the main cause of keratitis and viral encephalitis in developed countries.Materials and Methods:
Composite primers for PCR amplification of Leishmania major kDNA (kinetoplast DNA) were designed and optimized to use as IAC-HSV. IAC-HSV amplified in a non-stringent condition, ligated into pTZ57R plasmid vector, and transformed into Escherichia coli JM207 and then cloned. Resulting IAC was used for 105 CSF and 78 keratitis specimens.Results:
PCR amplicons for HSV and IAC-HSV were 454-bp and 662-bp, respectively. Detection limit of IAC was determined as 1000 plasmids per PCR reaction. IAC sensitivity for HSV detection was determined as 1000 plasmids per PCR reaction. IAC sensitivity for HSV detection was 500 copies/mL of HSV DNA. Among all specimens, 7 inhibited specimens were detected.Conclusions:
Indeed, using other DNA as an IAC is expected to detect false-negative results and amplification of the DNA is the key tool to examine the accuracy of amplification and detection steps. This internal amplification control is applicable for early reliable diagnosis of HSV in different loads of virus in different specimens.Keywords
1. Background
Herpes simplex virus (HSV) is responsible for a variety of serious diseases such as infection of cornea (keratitis) leading to corneal blindness and perilous infection of central nervous system such as encephalitis, which is the most common form of sporadic viral encephalitis in western countries (1, 2). The above-mentioned issues provide extreme necessity for a rapid, reliable, and specific diagnostic approach. Therefore, a suitable method of detection should provide the results in the shortest possible time and highest accuracy to inform the respective physician or patient (3).
Although viral culture is the gold standard for detection of HSV DNA, PCR techniques are more sensitive and the results are available in less than 4 hours (4). Recently, PCR has been developed extensively for virus infection diagnosis by many laboratories. This technique has beneficial characteristics including reliability, speed, and sensitivity that are vital for an ideal diagnostic assay. In spite of its outstanding advantages, PCR has some problems such as false-positive and -negative results (5). Clearly, by using a negative control (external control) during the PCR assay a false-positive result can be resolved. However, the major drawback of PCR is in producing false-negative results that remain unresolved. False-negative results are obtained from the malfunction of thermal cycler, imprecise extraction, insufficient Taq DNA polymerase, and other inhibitory substances that the exact act of these inhibitors are largely unknown (6). Although several extraction protocols and commercial kits have been developed to remove the probable PCR inhibiting substances from the specimen (7) and they seem to be ineffective for inhibitory removal that leads to a decrease of PCR sensitivity.
In order to enhance the certainty in negative PCR results, the presence of internal amplification control (IAC) is required (8, 9). IAC is a DNA sequence that is added to the sample and will be amplified with the target DNA simultaneously. Although IAC is commonly used for the detection of availability of PCR inhibitors (10), few technical problems were observed during IAC construction. Here a simple method for IAC construction with the least practical difficulties was used because of the above-mentioned requirements. Therefore, utilizing an IAC DNA in each PCR reaction that simultaneously amplified with viral target DNA with the same thermal profile used as an effective tool to overcome false-negative results in the PCR diagnosis of HSV.
Few studies have been carried out on the subject of HSV internal control. Burggraf and Olgemoller (11) constructed a single-stranded oligonucleotide as an internal control for a newly designed real time PCR for rapid detection of HSV. The amplified internal control was detected by the same probe that detects the amplified target. In another study, heterologous DNA was used to establish a multiple internal control DNA contained five IAC for five competitive Light Cycler-PCR assays including HSV IAC by using hybridization probes (12). The current investigation develops a simple and accurate path for IAC construction that has never been used by real-time and conventional PCR for HSV diagnosis.
In this study, IAC was designed in a competitive way by employing composite primer technique. Leishmania major kDNA (kinetoplast DNA) (MRHO: SU: 59: Pstrain, 620 bp) (13) was selected and amplified with composite primers. The main reason for the selection of kDNA contains different annealing temperatures from 40–70°C and annealing time duration from 10–60 seconds, which makes it suitable for construction of IAC. Therefore, by using the constructed IAC, an accurate and reliable approach was designed to examine the PCR amplification steps for HSV diagnosis, which is a common routine assay for all diagnosis laboratories (14).
2. Objectives
The purpose of this study was to design a competitive IAC for PCR diagnosis of HSV, which is the main cause of keratitis and viral encephalitis in developed countries. This IAC can be easily used in any diagnosis laboratory.
3. Materials and Methods
3.1. Specimens
A total of 105 cerebrospinal fluid (CSF) samples from patients with central nervous system diseases (suspected of HSV, cytomegalovirus (CMV), Epstein-Barr virus (EBV), varicella zoster virus (VZV), and human herpes virus 6 (HHV-6) (15, 16) and 78 samples from corneal epithelium (suspected of HSV, EBV, and CMV) of patients with herpetic dendritic keratitis and uveitis (17, 18) were obtained from the Virology Department of the Pasteur Institute in Iran (Tehran) that were collected from different hospitals (unknown). Specimens were used for comparison tests and examinations of the performance of constructed IAC. CSF specimens were obtained by subsequent lumbar punctures and aqueous samples from uveitis were obtained by anterior chamber paracentesis. All samples were collected from 2010-2011 and then were stored at 4°C until processed.
3.2. Preparation of Genomic DNA
To extract DNA, a DNP™ Extraction Kit (CinnaGen, Iran) was used. Final pellets were then mixed with 30 µL of double distilled water and kept at 4°C until used. Genomic DNA (HSV-1, KOS strain) was obtained from the Pasteur Institute of Iran (Tehran) to use as a positive control. HSV-1 (KOS strain) was cultured using VERO cell line (National Cell Bank, Pasteur Institute of Iran, Tehran) on RPMI medium (Sigma Co, USA) supplemented with penicillin G (10000 U), streptomycin (100 µg), and 10% FBS in 1 L of RPMI medium for examination as a positive control. Subsequent to the observation of 70–90% cytopathic effects viruses were harvested and kept at 4°C until used.
3.3. Designing HSV Primer.
Amplification primers for HSV were designed online at Invitrogen website (http://www.invitrogen.com). Designed primers were specific for pol gene (454 bp, accession number: AB231460.1). Primer sequences (constructed by CinnaGen Company, Tehran, Iran) were as follows: F-HSV: 5'ACCTACCGGCATACAAGCTCA 3', R-HSV: 5'AAGTGGCTCTGGCCTATGTCC3'.
3.4. PCR Condition
The PCR reaction contained TE buffer 10 x Tris-HCl (10 mm) and 1 mM of EDTA (pH 8.0), 1 µL of each forward and reverse HSV primers, MgCl2 (50 mm), dNTPs (10 mm), 2 units Taq DNA polymerase (CinnaGen, Iran), and 5 µL of viral DNA template and 13.5 µL of double distilled water to obtain a final volume of 25 µL. Amplification was performed by using Bioneer thermal cycler (Korea). The thermal profile was as follows: initial denaturation at 94°C for 5 minutes, followed by denaturation at 93°C for 20 seconds, annealing, and extension at 70°C for 20 seconds (40 cycles), and final extension at 72°C for 5 minutes. PCR products were electrophoresed and visualized on 1.5% agarose (stained with SYBR Green, CinnaGen, Iran) and markers SM1108 (50-1500 bp, MBI Fermentas, USA) were used.
3.5. Sensitivity and Specificity of HSV
3.5.1. Sensitivity Testing
Serial dilutions of HSV DNA ranged from 1 million to 10 copies/mL (by real-time PCR) were prepared for PCR amplification as described above. In addition, positive and negative controls were included.
3.5.2. Specificity Testing
Other species of Herpesviridae genomic DNA, such as VZV, EBV, CMV, and non-herpes genomic DNA including, Hepatitis B virus Salmonella Spp Mycobacterium tuberculosis (all were obtained from Pasteur Institute of Iran, Tehran), human genome and HSV as positive control were amplified by PCR with HSV primers in described PCR condition to test PCR specificity for the target site of primers. For all samples, 5 µL of genome equivalents was added to each PCR mixture. PCR products were then visualized as previously mentioned.
3.6. IAC Construction: Design of Composite Primer
L. major kDNA (MRHO: SU: 59: P strain, 620 bp) was selected to construct IAC. Amplification primers, which are unique for conserved sequences of kDNA, are as follows; Forward: 5′ TCGCAGAACGCCCC ACC 3′ and reverse: 5′ AGGGGTTGGTGTAAAATAGGC3′ (13). Forward and reverse primer sequences of HSV that were followed by forward and reverse primer sequences of kDNA are as follows: F-IAC-HSV: 5’ACCTACCGGCATACAAGC TCA TCGCAGAACGCCCCTACC 3’ and R-IAC-HSV: 5’ AAGTGGCTCTGGCCTATGTCCAGGGGT TGGTGTAAAATAGGC3’. Produced composite primer (Cinnagen, Iran) was used to amplify the 620 bp fragment (L. major) by using PCR with the following heat profile: denaturation at 30°C for 40 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 30 seconds (40 cycles), and a final extension at 72°C for 10 minutes. Obviously, constructed composite primer was only used to make IAC and for final detection of target DNA, only HSV primers used.
3.7. Cloning
Prior to the cloning process, PCR products were purified to obtain better results. To clone IAC fragments (662 bp) into the plasmid vector pTZ57R (2886 bp), the InsTAclone™ PCR Cloning Kit (cat: K1214, Fermentas, USA,) was employed. Briefly, 3 µL vector, 4 µL insert DNA, 1 µL T4 DNA ligase (5u), and 6 µL 5 x ligation buffer were mixed at a 30 µL final volume and left at room temperature for 4 hours. The physical process has done, as well. E. coli JM107 (Fermentas, USA) was competent by heat-shock and calcium chloride method as host cell and incubated at 37°C for 24 hours to improve the penetration of plasmid into the host. Then, 10 µL of ligation mixture containing recombinant plasmid pIACHSV (3,548 bp) was transformed into 50 µL of competent E. coli. Afterward, the suspension was cultured on Luria Bertani agar (Merck, Germany) containing ampicillin (100 µg/mL, Razi Co., Iran) and X-Gal and IPTG (2 µL/mL, Fermentas, USA). The resulting substances were then incubated at 37°C for 24 hours and alpha-complementation method was used to discover the recombinant colony containing pIACHSV (19).
3.8. Plasmid Extraction
Colonies containing inserted plasmid (pIACHSV) were extracted and purified by an Accuprep® Plasmid Extraction Kit (Cat. No. 3112, Pioneer, Korea) according to the manufacturer’s instructions and the extracted plasmid was kept at 4°C.
3.9. Determination of Standard Concentration of IAC
To obtain an ideal IAC concentration for each PCR reaction, serial 10-fold dilution of IAC (10-1 to 10-6) was amplified by PCR in combination with 1000 copies/mL of HSV DNA in each reaction tube with previously described reagents and thermal profile. Primary amount of IAC was measured by a NanoDrop spectrophotometer, A260 nm, Eppendorf, Germany. PCR products were visualized as described above.
3.10. Co-Amplification of HSV and IAC
A series of diluted HSV DNA ranging from 1 million to 500 copies/mL were prepared to observe the simultaneous co-amplification of pIACHSV and HSV DNA and to test the sensitivity of IAC in the presence of target DNA. In addition to other PCR reagents, each reaction was contained 1 µL of IAC in a final volume of 25 µL. For more confirmation of IAC, the PCR assay was performed for all specimens with an HSV thermal profile as described above.
3.11. Comparative Detection: PCR and Culture
A total of 105 CSF and 78 keratitis specimens were obtained from the Virology Department of Pasteur Institute in Iran (Tehran). Each specimen (0.25 mL) was incubated in ice for 1 hour prior to the culture. VERO cells (National Cell Bank, Pasteur Institute of Iran, Tehran) were added to RPMI medium (Sigma Co., USA), which was supplemented with penicillin G (10000 U), streptomycin (100 µg), and 10% FBS. The specimens were inoculated onto the flask of VERO cells and then incubated at 37°C. Cultures were then examined for cytopathic effects (70-90%) after 72 hours. Finally, PCR analysis for each specimen with IAC included negative and positive controls were performed as a referee to confirm the results (20).
4. Results
4.1. Sensitivity and Specificity test
A 10-fold serial dilution of HSV genomic DNA was used to find the range of sensitivity in PCR assay. Expected 454 bp fragments were observed on the gel. The PCR was able to detect a range between 1 million to 10 copies/mL of HSV DNA (Figure 1). It can be seen that PCR is able to detect the lowest concentrations of HSV (10 copies/mL), which implies the high sensitivity of PCR assay in HSV detection. Genomic DNA of other species of HSV and non-HSV DNA were used to test the specificity of the HSV PCR assay. Figure 2 shows no signal appeared due to the absence of homologous target sequences (All data not shown).
Sensitivity of PCR to Different Loads of HSV DNA

Specificity of HSV PCR

4.2. IAC Detection
kDNA (620 bp) of L. major was used for amplification with designed composite primers to produce 662 bp DNA as IAC. The resulting 662 bp fragments of DNA were cloned and transformed into a host and the new recombinant plasmids, which are known as pIACHSV, were then extracted. Then by using blue/white screening (alpha-complementation), few white colonies were isolated and their new recombinant plasmids, which are known as pIACHSV, were extracted. The expecting 662 bp signal appeared after running it on the gel.
4.3. IAC Optimization
After obtaining the expected 662 bp PCR amplicon, the ideal concentration of IAC was optimized and there could be no inhibition of amplification of target DNA. PCR amplification of IAC was examined by using different loads of IAC DNA. As a result, the lowest responsible concentration of IAC that could produce a distinct signal on the gel was 1000 plasmids (NanoDrop spectrophotometer, A260 nm, Eppendorf, Germany), which is preferred as an ideal concentration of IAC to be used per reaction (Figure 3).
Simultaneous Amplification of HSV DNA and IAC for Limit Detection

4.4. Detection of HSV in the Presence of IAC
The sensitivity of PCR detection for HSV in the presence of IAC was examined by mixing different concentrations of the HSV template in the presence of IAC DNA. Two expecting signals for HSV and IAC DNA were observed at 454 bp and 662 bp, respectively. The highest sensitivity of IAC and HSV co-amplification were obtained at 500 copies/mL for HSV DNA. The concentration of IAC remained constant at 1000 plasmids within a broad range of HSV DNA concentrations without the affection of the target signal. However, the amplification of target DNA disappeared at 50 copies/mL. The target DNA amplification was inhibited when less than 500 copies/mL of the target template was used in the reaction.
4.5. Comparison of PCR and Culture
A total of 105 CSF and 78 keratitis specimens were examined by both culture and PCR assay to demonstrate the comparative diagnostic results. It can be seen from the CSF specimens that the PCR assay generated 21 positive results; however, in the culture method, only 10 positive results had been achieved. In the case of the keratitis specimens, 15 positive results were achieved by PCR and 10 by culture. PCR was highly sensitive (100%) when it was used as a reference method. Therefore, the sensitivity of PCR and culture in the case of CSF specimens were 100% and 27.6%, respectively. However, the sensitivity of PCR and culture in the case of keratitis specimens were 100% and 66.66%, respectively. In addition, the specificity was 100% for both methods.
4.6. Analyzing of Co-amplification of Specimens Genomic DNA and IAC
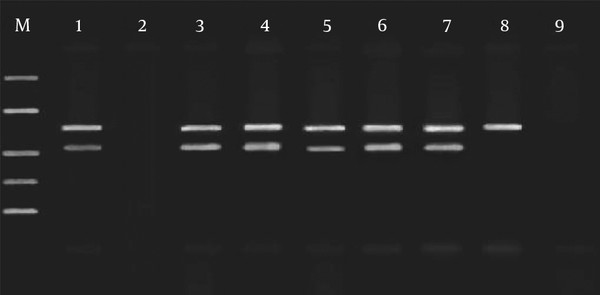
PCR assay was carried out for CSF and keratitis specimens in the presence of IAC to confirm the exact function and performance of IAC. Figure 4 shows there is an inhibited specimen, which indicates that one or more factors have not functioned correctly in the course of extortion or any other possible problems. In addition, it can be seen that there is a true-negative results for which only the IAC signal was produced. In the current experiments, no unexpected results were observed. However, in the case of presence of a very low amount of target DNA, no signal may be seen in the results. Table 1 shows all data related to the inhibited and uninhibited results.
Simultaneous Amplification of HSV DNA and IAC of Some CSF and Keratitis Specimens, Representing 7 Experiments, 4 for CSF and 3 for Keratitis
| Sample Results | Inhibited | Uninhibited |
|---|---|---|
| Positive | 3 | 33 |
| Negative | 4 | 143 |
| Total | 7 | 176 |
5. Discussion
In our investigation, the specificity of the primer set was examined against other species of Herpesviridae and non-HSV genomic DNA. Although most of the employed DNA in the specificity test was DNA HSV-dependent, still no signal was observed that shows the precision and specificity in primer sequences (Figure 2). In this study, sensitivity testing of the PCR assay showed that this method is able to detect at least 10 copies/mL of HSV DNA. Thus, it can be concluded that this technique has high efficiency for HSV diagnosis. An IAC is a non-target DNA sequence, which is present in the tube reaction along with HSV DNA that will be amplified simultaneously in the PCR assay under the same condition. It should be mentioned that IAC should have unique characteristics such as stable, non-infectious, easily produced, and standardized (4).
As already mentioned in the previous section, there are two major approaches for IAC construction competitive and noncompetitive approach (21). In a noncompetitive approach, there are two different set of primers for IAC and target DNA. Therefore, two PCR reactions will be carried out in one PCR tube with the same thermal profile. The optimization of IAC with this method seems to be complicated due to the extensive range of annealing temperatures for IAC and target as well as the increase of risk to produce nonspecific signals due to the use of two different set of primers (22). On the other hand, the competitive method has only one set of primers for IAC and target DNA. Thus, there is constant competition between IAC and target DNA for binding to the primers. The main drawback of this method is the decrease of sensitivity of PCR due to the enhancement or reduction of amplification in one of the products (IAC or target DNA) that can lead to a false-negative result. Thus, the most critical factor to overcome this issue is to determine the proper concentration of IAC. In this way, IAC is able to compete with the DNA target to bind with the primers reasonably (23). In a sample reaction, a high concentration of IAC is not able to detect weak inhibitors. However, even low concentrations of these inhibitors can cause the target DNA signal to disappear (especially in low concentration of target DNA). On the other hand, when the amount of target DNA is high, a low concentration of IAC will be eliminated due to the presence of competition during the amplification process (14). Here, 1000 plasmids were determined as an ideal concentration of IAC in each reaction. This amount is sensitive for concentrations higher than 500 copies/mL of target DNA.
In this work, a novel IAC was constructed by means of an easy and applicable approach. The kDNA of L. major was used for construction of IAC by the composite primer technique. Through such methods, only one set of primer was used during the PCR amplification. Composite primers were made by introducing HSV primer sequences to the 5’ ends of kDNA primer sequences. The resulting composite primers have 5’ over-hanging ends that are complementary to HSV primers. The selected primers for IAC are relatively short, which eliminate the risk of obtaining nonspecific bands on the gel (Figures 3 and 4). The employed gene for IAC in our study has a broad spectrum annealing temperature and, as a result, is insensitive to thermal profile (14). This means that such an optimization is not complicated. Furthermore, utilizing a cloning technique provides a limitless source of IAC as a recombinant DNA that can be easily used in diagnostic laboratories. Moreover, the difference in size of IAC fragments and HSV DNA is suitable and easy to detect. It is necessary to know that the presence of two sets of primer sequences in one reaction will increase the possibility of nonspecific band production. Therefore, we used the composite primer technique to have only one set.
When the target signal was observed, it meant that the inhibited IAC amplification has occurred due to the high concentration of target DNA. Consequently, with the amplification of the target DNA, the amplification of IAC is not mandatory. Thus, a positive result is considered true. If neither IAC nor the target DNA have generated a signal, it can be concluded that at least one inhibitor was present in the PCR assay (24). At this stage, all of the steps have to be repeated properly. The obtained results from the culture method indicated that PCR is a much more sensitive technique for HSV diagnosis in comparison with the viral culture method and would be considered a reference technique (25, 26) regardless of whether the samples were obtained from CSF or keratitis.
In the recent years, IAC was constructed for various pathogens by means of different DNAs (27, 28). In addition, various genes can be used for an IAC. For example, Dreier et al. (22) used λ Bacteriophage to make an IAC for validation of detecting congenital Cytomegalovirus. This IAC was 150 bp and target gene was 100 bp. The gap between the two fragments (IAC and target DNA) should be relatively high and be easily distinguishable (28). In the present investigation, the size difference in the constructed IAC fragments and HSV DNA is about 200 bp, which can be easily distinguished. Also, in different studies (23, 29), IAC were constructed for real-time PCR diagnosis for different pathogens by using single stranded oligonucleotides. However, we designed an IAC for general PCR and by using non-target gene (kDNA f Leishmania), which would be used for construction of IAC for many other target pathogens that PCR diagnosis was due to a broad annealing temperature. By using introduced IAC, which is applicable easily with broad range of annealing temperature, to establish a new reliable method to have the least false-negative results for this kind of diagnosis.
The methodology is simple and should enable laboratories to conveniently complement their existing panel of PCR assays with such an IAC. Until now, the concentration of IAC and target sequence was considered a critical parameter for the detection limit of a PCR assay. The demonstrated stability in the current study makes this DNA suitable for the construction of IAC in other pathogenic DNA, a fact that can also be applicable for other molecular based viral detection systems. Moreover, the higher amount of target DNA present in the samples, the lower was the increase of the IAC specific amplification signals on the gel. As a result, during the complementing of PCR assay with competitive IAC, the amount of IAC plays an important role that can avoid the competitive inhibition of the respective virus-specific PCR and loss of assay sensitivity. It can be seen that due to the competition between the two DNAs in the sample reaction, a high concentration of IAC in the sample of reaction is not influenced by feeble and least inhibitors. However, the disappearance of the target band by those weak inhibitors is possible. Therefore, determination of an optimized amount of IAC in each PCR reaction is vital (21, 30, 31) that, in fact, can be a limitation of this study that can be improved with further investigation. Since the introduced IAC may not express all inhibitory factors in HSV diagnosis and it might just explain some of them.
In conclusion, if a nucleic acid amplification method is to be a highly reliable diagnostic test for pathogens, it must include internal control. Here, a novel IAC with a competitive approach was constructed for HSV diagnosis, which was in an agreement and not a reflection with the HSV thermal profile. It can be a sensitive tool to obtain reliable results for HSV diagnosis with a PCR assay, which is a cost effective, simple, and accurate method. It can be applicable in routine evaluations or diagnostic kits. The use of IAC DNA may streamline assays and may avoid the risk of handling errors in HSV detection due to the addition of only one IAC to the various samples.
Acknowledgements
References
-
1.
Pruss H, Finke C, Holtje M, Hofmann J, Klingbeil C, Probst C, et al. N-methyl-D-aspartate receptor antibodies in herpes simplex encephalitis. Ann Neurol. 2012;72(6):902-11. [PubMed ID: 23280840]. https://doi.org/10.1002/ana.23689.
-
2.
Wang JP, Bowen GN, Zhou S, Cerny A, Zacharia A, Knipe DM, et al. Role of specific innate immune responses in herpes simplex virus infection of the central nervous system. J Virol. 2012;86(4):2273-81. [PubMed ID: 22171256]. https://doi.org/10.1128/JVI.06010-11.
-
3.
Pena KC, Adelson ME, Mordechai E, Blaho JA. Genital herpes simplex virus type 1 in women: detection in cervicovaginal specimens from gynecological practices in the United States. J Clin Microbiol. 2010;48(1):150-3. [PubMed ID: 19923487]. https://doi.org/10.1128/JCM.01336-09.
-
4.
Oberhelman RA, Soto-Castellares G, Gilman RH, Caviedes L, Castillo ME, Kolevic L, et al. Diagnostic approaches for paediatric tuberculosis by use of different specimen types, culture methods, and PCR: a prospective case-control study. Lancet Infect Dis. 2010;10(9):612-20. [PubMed ID: 20656559]. https://doi.org/10.1016/S1473-3099(10)70141-9.
-
5.
Burkardt HJ. Standardization and quality control of PCR analyses. Clin Chem Lab Med. 2000;38(2):87-91. [PubMed ID: 10834394]. https://doi.org/10.1515/CCLM.2000.014.
-
6.
Cortez-Herrera E, Sperhacke R, Becker D, Kritski A, Zaha A, Rossetti MR. Internal control in pcr for mycobacterium tuberculosis: Usefulness and improvement of the diagnosis. Braz Arch Biol Technol. 2008;51(4):485-91. https://doi.org/10.1590/s1516-89132008000400006.
-
7.
Escobar-Herrera J, Cancio C, Guzman GI, Villegas-Sepulveda N, Estrada-Garcia T, Garcia-Lozano H, et al. Construction of an internal RT-PCR standard control for the detection of human caliciviruses in stool. J Virol Methods. 2006;137(2):334-8. [PubMed ID: 16857274]. https://doi.org/10.1016/j.jviromet.2006.06.015.
-
8.
Bosch A, Sánchez G. Analytical methods for virus detection in water and food. Food Anal Method. 2011;4(1):4-12.
-
9.
Rodriguez-Lazaro D, Hernández M. Real-time PCR in food science: introduction. Curr Iss Mol Biol. 2013;15(2):25-38.
-
10.
Barkham T, Hoorfar J. Internal amplification control for PCR should not be mandatory in the clinical medical environment. J Clin Microbiol. 2004;42(7):3379-80. [PubMed ID: 15243125]. https://doi.org/10.1128/JCM.42.7.3379-3380.2004.
-
11.
Burggraf S, Olgemoller B. Simple technique for internal control of real-time amplification assays. Clin Chem. 2004;50(5):819-25. [PubMed ID: 15010426]. https://doi.org/10.1373/clinchem.2003.027961.
-
12.
Stocher M, Leb V, Berg J. A convenient approach to the generation of multiple internal control DNA for a panel of real-time PCR assays. J Virol Methods. 2003;108(1):1-8. [PubMed ID: 12565148].
-
13.
Mahboudi F, Abolhassani M, Tehrani SR, Azimi M, Asmar M. Differentiation of old and new world leishmania species at complex and species levels by PCR. Scand J Infect Dis. 2002;34(10):756-8. [PubMed ID: 12477327].
-
14.
Lund M, Madsen M. Strategies for the inclusion of an internal amplification control in conventional and real time PCR detection of Campylobacter spp. in chicken fecal samples. Mol Cell Probes. 2006;20(2):92-9. [PubMed ID: 16330185]. https://doi.org/10.1016/j.mcp.2005.10.002.
-
15.
Minjolle S, Arvieux C, Gautier AL, Jusselin I, Thomas R, Michelet C, et al. Detection of herpesvirus genomes by polymerase chain reaction in cerebrospinal fluid and clinical findings. J Clin Virol. 2002;25 Suppl 1:S59-70. [PubMed ID: 12091083].
-
16.
Gaeta A, Verzaro S, Cristina LM, Mancini C, Nazzari C. Diagnosis of neurological herpesvirus infections: real time PCR in cerebral spinal fluid analysis. New Microbiol. 2009;32(4):333-40. [PubMed ID: 20128439].
-
17.
Crouse CA, Pflugfelder SC, Pereira I, Cleary T, Rabinowitz S, Atherton SS. Detection of herpes viral genomes in normal and diseased corneal epithelium. Curr Eye Res. 1990;9(6):569-81. [PubMed ID: 2167192].
-
18.
Sugita S, Shimizu N, Watanabe K, Ogawa M, Maruyama K, Usui N, et al. Virological analysis in patients with human herpes virus 6-associated ocular inflammatory disorders. Invest Ophthalmol Vis Sci. 2012;53(8):4692-8. [PubMed ID: 22700707]. https://doi.org/10.1167/iovs.12-10095.
-
19.
Moslemi E, Shahhosseiny MH, Javadi G, Praivar K, Sattari TN, Amini HK. Loop mediated isothermal amplification (LAMP) for rapid detection of HBV in Iran. Afr J Microbiol Res. 2009;3(8):439-45.
-
20.
Singh A, Preiksaitis J, Ferenczy A, Romanowski B. The laboratory diagnosis of herpes simplex virus infections. Can J Infect Dis Med Microbiol. 2005;16(2):92-8. [PubMed ID: 18159535].
-
21.
Sachadyn P, Kur J. The construction and use of a PCR internal control. Mol Cell Probes. 1998;12(5):259-62. [PubMed ID: 9778450]. https://doi.org/10.1006/mcpr.1998.0170.
-
22.
Dreier J, Stormer M, Kleesiek K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J Clin Microbiol. 2005;43(9):4551-7. [PubMed ID: 16145106]. https://doi.org/10.1128/JCM.43.9.4551-4557.2005.
-
23.
Das A, Spackman E, Senne D, Pedersen J, Suarez DL. Development of an internal positive control for rapid diagnosis of avian influenza virus infections by real-time reverse transcription-PCR with lyophilized reagents. J Clin Microbiol. 2006;44(9):3065-73. [PubMed ID: 16954228]. https://doi.org/10.1128/JCM.00639-06.
-
24.
Hoorfar J, Malorny B, Abdulmawjood A, Cook N, Wagner M, Fach P. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J Clin Microbiol. 2004;42(5):1863-8. [PubMed ID: 15131141].
-
25.
Thompson PP, Kowalski RP. A 13-year retrospective review of polymerase chain reaction testing for infectious agents from ocular samples. Ophthalmology. 2011;118(7):1449-53. [PubMed ID: 21367461]. https://doi.org/10.1016/j.ophtha.2010.12.004.
-
26.
Ziyaeyan M, Alborzi A, Jamalidous M, Japoni A, Badiee P, Moeini M. Diagnosis and Quantitative Detection of Herpes Simplex Virus DNA in Corneal Ulcers. Int J Virol. 2012;8(3):279-84. https://doi.org/10.3923/ijv.2012.279.284.
-
27.
Nolte FS. Novel internal controls for real-time PCR assays. Clin Chem. 2004;50(5):801-2. [PubMed ID: 15105347]. https://doi.org/10.1373/clinchem.2004.032672.
-
28.
Jones RN, Neale ML, Beattie B, Westmoreland D, Fox JD. Development and application of a PCR-based method including an internal control for diagnosis of congenital cytomegalovirus infection. J Clin Microbiol. 2000;38(1):1-6. [PubMed ID: 10618053].
-
29.
Chen J, Zhang L, Paoli GC, Shi C, Tu SI, Shi X. A real-time PCR method for the detection of Salmonella enterica from food using a target sequence identified by comparative genomic analysis. Int J Food Microbiol. 2010;137(2-3):168-74. [PubMed ID: 20060189]. https://doi.org/10.1016/j.ijfoodmicro.2009.12.004.
-
30.
de Vries JJ, van der Eijk AA, Wolthers KC, Rusman LG, Pas SD, Molenkamp R, et al. Real-time PCR versus viral culture on urine as a gold standard in the diagnosis of congenital cytomegalovirus infection. J Clin Virol. 2012;53(2):167-70. [PubMed ID: 22177273]. https://doi.org/10.1016/j.jcv.2011.11.006.
-
31.
Oikonomou I, Halatsi K, Kyriacou A. Selective PCR: a novel internal amplification control strategy for enhanced sensitivity in Salmonella diagnosis. Lett Appl Microbiol. 2008;46(4):456-61. [PubMed ID: 18298457]. https://doi.org/10.1111/j.1472-765X.2008.02340.x.