Abstract
Background:
Petroleum reservoirs have long been known as the hosts of extremophilic microorganisms. Some of these microorganisms are known for their potential biotechnological applications, particularly production of extra and intracellular polymers and enzymes.Objectives:
Here, 14 petroleum liquid samples from southern Iranian oil reservoirs were screened for presence of biosurfactant‐producing halothermophiles.Materials and Methods:
Mixture of the reservoir fluid samples with a minimal growth medium was incubated under an N2 atmosphere in 40°C; 0.5 mL samples were transferred from the aqueous phase to agar plates after 72 hours of incubation; 100 mL cell cultures were prepared using the MSS-1 (mineral salt solution 1) liquid medium with 5% (w/v) NaCl. The time-course samples were analyzed by recording the absorbance at 600 nm using a spectrophotometer. Incubation was carried out in 40°C with mild shaking in aerobic conditions. Thermotolerance was evaluated by growing the isolates at 40, 50, 60 and 70°C with varying NaCl concentrations of 5% and 10% (w/v). Halotolerance was evaluated using NaCl concentrations of 5%, 10%, 12.5% and 15% (w/v) and incubating them at 40°C under aerobic and anaerobic conditions. Different phenotypic characteristics were evaluated, as outlined in Bergey's manual of determinative bacteriology. Comparing 16S rDNA sequences is one of the most powerful tools for classification of microorganisms.Results:
Among 34 isolates, 10 demonstrated biosurfactant production and growth at temperatures between 40°C and 70°C in saline media containing 5%‐15% w/v NaCl. Using partial 16S rDNA sequencing (and amplified ribosomal DNA restriction analysis [ARDRA]) and biochemical tests (API tests 20E and 50 CHB), all the 10 isolates proved to be facultative anaerobic, Gram-positive moderate thermohalophiles of the genus Bacillus (B. thermoglucosidasius, B. thermodenitrificans, B. thermoleovorans, B. stearothermophilus and B. licheniformis), exhibiting surface-active behaviors.Conclusions:
General patterns include decreasing the thermotolerance with increasing the salt concentrations and also more halotolerance in the aerobic environment compared with anaerobic conditions. The results demonstrated that Iranian petroleum reservoirs enjoy a source of indigenous extremophilic microorganisms with potential applications in microbial enhanced oil recovery and commercial enzyme production.Keywords
1. Background
Research into the microbial life inhabiting the subterranean environments was traditionally important in biogeosciences, where it was crucial for understanding and explaining the chemistry and ecology of such systems. More recently, such knowledge has been useful in a number of engineered systems involving deep underground environments. A good example is oil reservoirs, considered for microbial enhanced oil recovery (MEOR) projects (1). Insight into the microbial community nature of a reservoir can be crucial in determining the suitability of the reservoir for an MEOR project, by providing information about the presence or absence of favorable species versus the unfavorable ones (e.g. sulfur-reducing bacteria) (2). Furthermore, such information contributes to understanding the possible mechanisms of enhanced oil recovery (interfacial tension reduction, selective plugging, etc.).
This in turn can be used in generating models that predict ultimate enhanced recovery, and optimizing the chemistry of injection fluids to promote a certain mechanism (e.g. biosurfactant production). Alternatively, lack of a potent microbial community can lead engineers to introduce exogenous species (3). Similar to other extreme environments, petroleum reservoirs typically host extremophilic organisms like haloalkaliphiles and thermohalophiles; a diverse group of microorganisms, characterized by their haloadaptive capabilities, enabling them to survive in hypersaline environments such as salt lakes, saline soils, salt mines, oil reservoirs, etc. (4-6). These microorganisms are generally categorized in two groups of extreme and moderate halophiles.
Extreme halophiles grow optimally in NaCl concentrations of 15%-30% (w/v) and generally belong to archaea, while moderate halophiles tolerate NaCl concentrations of 3%-15% (w/v) and are dominated by Bacteria (7). Moderate halophiles are better known for their potential biotechnological applications, particularly production of extra and intracellular polymers and enzymes (2). Enzymes produced by moderate halophiles have been the subject of extensive research because of their unique ability to perform optimally in high salt concentrations, which potentially eliminates the need for extensive sterilization procedures.
Amylase (8-13), esterase (12, 13), protease (5, 14-16), leucine dehydrogenase (17) and other halotolerant enzymes (18) have been successfully purified from moderate halophiles. In addition to extracellular enzymes, production of extracellular biosurfactants and bioemulsifiers has been reported by some moderate halophiles (1, 19, 20). Producing biosurfactant is favorable in the MEOR processes. In the present study, we reported isolation and characterization of biosurfactant-producing moderate halophiles from southern Iranian petroleum reservoirs. The results of this work demonstrated the potential of these reservoirs for MEOR and provided further information about their indigenous microbial populations.
2. Objectives
The aim of this study was isolation of biosurfactant-producing microorganisms from petroleum reservoirs with potential applications in microbially enhanced oil recovery and commercial enzyme production.
3. Materials and Methods
3.1. Samples
Reservoir fluid samples from southwestern Iranian oil reservoirs were collected from many well head sampling lines. Oil was allowed to flow from the sampling valve for 15 minutes prior to sampling. During the sampling, a part of the dissolved gas was allowed to bubble up from oil to force out the air in the container. To prevent environmental contamination, the remaining dissolved gas was used to keep the container slightly pressurized. Samples were then delivered to a laboratory and kept in ambient conditions.
3.2. Selective Isolation of Biosurfactant-Producing Microorganisms
Of each oil sample, 10 mL was mixed with 90 mL of a minimal growth medium containing 2.0 g KH2PO4, 5.0 g K2HPO4, 10 g NaCl, 3.0 g (NH4)2SO4, 0.002 g MnSO4.H2O, 0.2 g MgSO4.7H2O, 0.01 g CaCl2.2H2O, 10 g Glucose, and 1.0 g yeast extract per liter (all from Merck, Germany). The mixture was incubated under 100% N2 atmosphere (anaerobic condition) in 40°C. The 0.5 mL samples were transferred from the aqueous phase to agar plates after 72 hours of incubation. Individual colonies were isolated by further streaking. Production of extracellular biosurfactants by each isolate was evaluated by oil Spreading and emulsification (E24) tests, as described elsewhere (3, 21). Liquid cultures were prepared using the medium introduced by Folmsbee et al. (22) with the NaCl concentration adjusted to 5% (w/v). This medium was referred to as MSS-1 (mineral salt solution 1).
3.3. Evaluation of Growth and Sensitivity to Thermal and Saline Stresses
Cell cultures of 100 mL were prepared using the MSS-1 liquid medium with 5% (w/v) NaCl. Time-course samples were analyzed by recording the absorbance at 600 nm using a spectrophotometer (Optima, Japan). Incubation was carried out in 40°C with mild shaking in aerobic and anaerobic conditions. Thermotolerance was evaluated by growing the isolates at 40°C, 50°C, 60°C and 70°C with varying NaCl concentrations of 5% and 10% (w/v). Halotolerance was evaluated using NaCl concentrations of 5%, 10%, 12.5% and 15% (w/v), and incubating them at 40°C under aerobic and anaerobic conditions.
3.4. Phenotypic Characterization and 16S rDNA Analysis
Different phenotypic characteristics were evaluated, as outlined in Bergey's manual of determinative bacteriology (23), such as motility, Gram reaction, oxidation/fermentation of glucose, starch, esculin and gelatin hydrolyses, oxidase, catalase and lecithinase productions, citrate, xylose, arabinose and ribose utilizations and sensitivity to penicillin. Comparing the 16S rDNA sequences is one of the most powerful tools for classification of microorganisms (20). A group-specific primer pair capable of amplifying a specific sequence of 16S rDNA from all Bacillus taxa has been developed by Wu et al. (24): B-K1/F (5’-TCACCAAGGCRACGATGCG-3’) and B-K1/R1 (5’-CGTATTCACCGCGGCATG-3’) (MWG, Germany). Possible genomic differences between the isolates based on the amplicons generated by these primers, were evaluated by amplified ribosomal DNA restriction analysis (ARDRA).
The bacterial genomic DNA was isolated according to the modified method of Mantynen and Lindstrom (25). The purified genomic DNA was qualified by gel electrophoresis and its quantity was determined by recording the absorbance in 260 and 280 nanometers, using an ND-1000 Nanodrop (USA). For PCR experiments, a 50 µL solution of DNA with concentration of 100 ng/µL was prepared. Amplification was conducted with a 20 µL reaction volume containing 2 µL of 10X PCR buffer (TaKaRa, Japan), 0.1 mM dNTP mixture (TaKaRa, Japan), 10 pmol of each primer (MWG, Germany), 0.5 U of Taq DNA polymerase (TaKaRa, Japan) and 100 ng of total genomic DNA. Amplification was carried out in a TECHNE thermocycler (UK) under the following conditions: five minutes at 94°C, followed by 25 cycles of DNA amplification (30 seconds at 94°C, 30 seconds at 63°C and 120 seconds at 72°C) and a final extension at 72°C for 10 minutes. All amplification reactions were repeated two times under identical conditions accompanied by a negative control. ARDRA analysis of PCR amplicons was carried out using Alu1 (AG’CT) and Taq1 (T’CGA) restriction enzymes (Fermentas, Germany) according to the manufacturer’s instructions. Restricted products were separated on 2% agarose gels and visualized by UV excitation PCR. The products were purified using the QIAGEN PCR purification kit (USA). Of the purified PCR product, 50 µL was sent to the Tech Dragon Limited Company (China) for sequencing. Similarity search and sequence analysis were carried out using the BLAST server of NCBI.
4. Results
Thirty four bacterial isolates were isolated from the liquid samples through the procedure previously explained, out of which, 10 produced extracellular surface active compounds through the oil spreading and E24 tests (Table 1). Biochemical and molecular assays suggested that the isolates most resembled B. thermoglucosidasius, B. thermodenitrificans, B. thermoleovorans, B. stearothermophilus and B. licheniformis, as presented in Table 1. The results were confirmed by API tests 20E and 50 CHB, performed using the commercially available kits (data are not shown).
Phenotypic Characteristics and Preliminary Assay of Biosurfactant Production by the Isolated Strains a
| Number of Isolate | OS Index, cm | E24% | Motility | Citrate | Lecithinase | Terminal Spore | Central Spore | Subterminal Spore | Penicillin | Starch Hydrolysis | Gram Reaction | Anaerobic | Oxidase | Catalase | Gelatin L | Esculin | Fluorescence | Ribose | Arabinose | Xylose |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 1.0 | 0.29 | + | - | + | + | + | + | - | + | + | + | + | + | + | - | - | + | + | + |
| 8 | 0.5 | 0.17 | + | + | + | + | + | + | - | + | + | + | + | + | + | + | - | - | - | + |
| 14 | 1.0 | 0.21 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 71 | 3.0 | 0.32 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 5 | 0.5 | 0.31 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 21 | 0.8 | 0.53 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 103 | 3.5 | 0.93 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 13 | 2.0 | 0.30 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 12 | 3.0 | 0.14 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
| 51 | 0.5 | 0.18 | + | - | + | + | + | + | - | + | + | + | + | + | + | + | - | + | + | + |
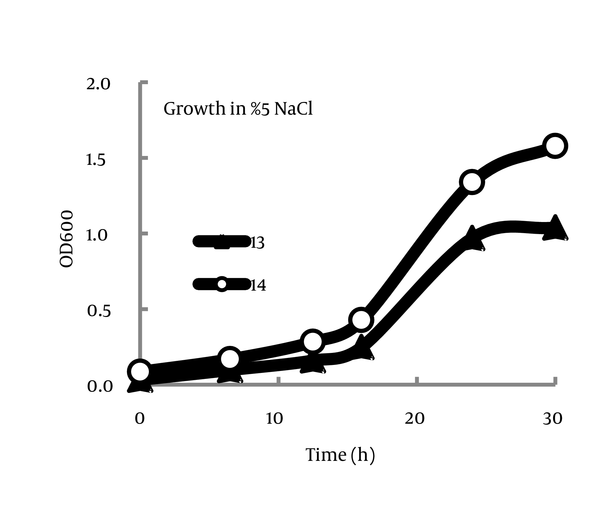
The isolates are fairly similar in growth kinetics in the studied conditions (Figure 1). As it is given in Tables 2 and 3, all the isolates demonstrated similar behaviors in varying saline and thermal conditions. General patterns included decreasing thermotolerance with increasing salt concentrations and also more halotolerance in the aerobic environment compared with anaerobic conditions.
The Sample Growth Curves for Isolates 13 and 14
Assessment of Thermotolerance in Salinities of 5% and 10% NaCl
| Number of Isolate | 5% NaCl | 10% NaCl | |||||
|---|---|---|---|---|---|---|---|
| 40°C | 50°C | 60°C | 70°C | 40°C | 50°C | 60°C | |
| 7 | + | + | + | - | + | + | - |
| 8 | + | + | + | - | - | - | - |
| 14 | + | + | + | - | + | + | - |
| 71 | + | + | + | - | - | - | - |
| 5 | + | + | + | - | + | + | - |
| 21 | + | + | + | - | + | + | - |
| 103 | + | + | + | - | - | - | - |
| 13 | + | + | + | - | - | - | - |
| 12 | + | + | + | - | + | + | - |
| 51 | + | + | + | + | - | - | - |
Assessment of Halotolerance in Aerobic and Anaerobic Conditions
| Number of Isolate | Anaerobic, 40°C | Aerobic, 40°C | ||||||
|---|---|---|---|---|---|---|---|---|
| 5% | 10% | 12.5% | 15% | 5% | 10% | 12.5% | 15% | |
| 7 | + | + | + | + | + | + | + | + |
| 8 | + | - | - | - | + | - | - | - |
| 14 | + | + | + | + | + | + | + | + |
| 71 | + | - | - | - | + | + | + | + |
| 5 | + | + | + | + | + | + | + | + |
| 21 | + | + | + | + | + | + | + | + |
| 103 | + | - | - | - | + | - | - | - |
| 13 | + | - | - | - | + | - | - | - |
| 12 | + | + | + | + | + | + | + | + |
| 51 | + | - | - | - | + | - | - | - |
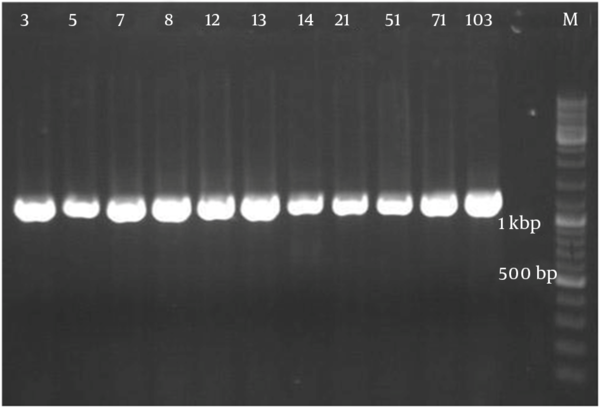
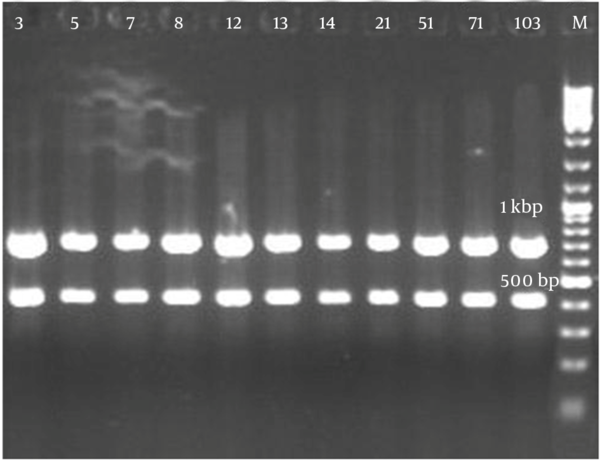
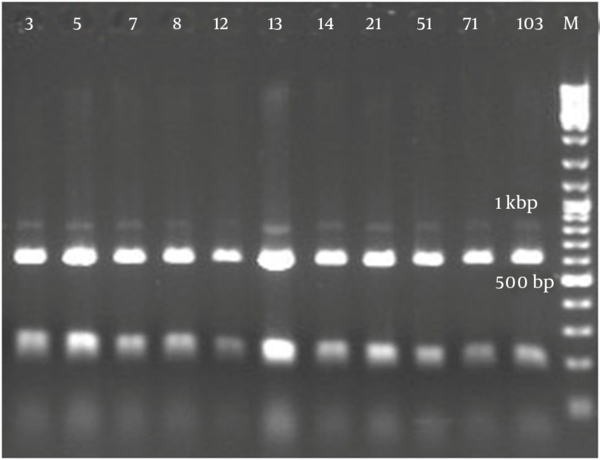
Figure 2 demonstrates that all the isolates of 1114 bp were specific to bacilli. Taq1 digested the PCR product identically and produced two amplicons with approximate sizes of 420 and 690 bp (Figure 3). Similarly, Alu1 digested the PCR product identically and produced five amplicons of 53, 59, 250, 252 and 600 bp (Figure 4) (the precise amplicon sizes were calculated using Vector NTI software, based on the sequencing data). Mse1 and Mbo1 were also tested, but failed to differentiate between the isolates. The similarity search using BLAST revealed over 99% homology with more than 1000 different bacilli.
Amplified Ribosomal DNA Restriction Analysis Using the Bacilli Group-Specific Primer Developed by Wu et al. (24)
Amplified Ribosomal DNA Restriction Analysis Using Taq1 (TCGA)
Amplified Ribosomal DNA Restriction Analysis Using Alu1 (AGCT)
5. Discussion
The results of 16S rDNA sequencing identical a 1114-bp sequence for these thermophilic bacilli. Therefore, implemented primers are not recommended for this class of bacilli. Wu et al. (24) successfully used this sequence to differentiate between varieties of bacilli including B. licheniformis, B. subtilis and B. ciculans. Manachini et al. (25) also reported that sequencing of 16s rDNA could not differentiate between Bacillus thermodenitrificans and their closest phylogenetic neighbors, e.g. the type strains of B. stearothermophilus and B. thermoleovorans, but they concluded that amplification of the internal transcribed spacer (ITS) region of rrn operons could significantly separate these strains (25). On the other hand, Vaerewijck et al. (26) reported that ARDRA using universal primers could not distinguish B. licheniformis and B. subtilis from B. pumilus and B. amyloliquefaciens. Kuisiene et al. (27), however, successfully identified the thermophilic strain Geobacillus stearothermophilus from similar strains, using ARDRA with Alu1 as their sequencing enzyme.
As given in Table 3, all the isolates demonstrated similar halotolerance behaviors in aerobic and anaerobic conditions, except for isolate 71, which did not growth under anaerobic conditions with salt concentrations more than 5%. It may be concluded that this isolate can only survive in low salt concentration reservoirs, or had a kind of symbiosis under high salt concentration conditions. The results of the present work contributed to the mounting evidence that Iranian petroleum reservoirs are host to a rich community of extremophiles. Previous studies demonstrated presence of Pseudomonas, Bacillus and Rhodococcus species in these areas and their successful implementation for MEOR in experimental core flooding setups (28-31). All of these studies focused specifically on biosurfactant-producing species. However, the results of isolation experiments suggest that microbial communities of these reservoirs are more diverse and might not be limited to bacteria.
Although, little is known about the obligate anaerobes in these reservoirs of which preliminary experiments suggest their existence (data are not shown). Future work can address various questions related to basic microbiology of the organisms, or focus on optimizing the parameters important to engineering their growth. The methods used in this study have been tailored for selective isolation of biosurfactant-producing extremophiles; however, similar experimental approaches can be used to study the microbial communities in broader microbiological contexts, to encompass nonbacterial organisms, extreme halophiles, and obligate anaerobes. Finally, potential applications of such organisms in enzyme production can be explored.
Acknowledgements
References
-
1.
Dastgheib SM, Amoozegar MA, Elahi E, Asad S, Banat IM. Bioemulsifier production by a halothermophilic Bacillus strain with potential applications in microbially enhanced oil recovery. Biotechnol Lett. 2008;30(2):263-70. [PubMed ID: 17876532]. https://doi.org/10.1007/s10529-007-9530-3.
-
2.
Quillaguaman J, Hashim S, Bento F, Mattiasson B, Hatti-Kaul R. Poly(beta-hydroxybutyrate) production by a moderate halophile, Halomonas boliviensis LC1 using starch hydrolysate as substrate. J Appl Microbiol. 2005;99(1):151-7. [PubMed ID: 15960675]. https://doi.org/10.1111/j.1365-2672.2005.02589.x.
-
3.
Youssef NH, Duncan KE, Nagle DP, Savage KN, Knapp RM, McInerney MJ. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004;56(3):339-47. [PubMed ID: 14967225]. https://doi.org/10.1016/j.mimet.2003.11.001.
-
4.
Gold T. The deep, hot biosphere. Proc Natl Acad Sci USA. 1992;89(13):6045-9. [PubMed ID: 1631089].
-
5.
Kato T, Haruki M, Imanaka T, Morikawa M, Kanaya S. Isolation and characterization of long-chain-alkane degrading Bacillus thermoleovorans from deep subterranean petroleum reservoirs. J Biosci Bioeng. 2001;91(1):64-70. [PubMed ID: 16232948].
-
6.
Ventosa A, Marquez MC, Garabito MJ, Arahal DR. Moderately halophilic gram-positive bacterial diversity in hypersaline environments. Extremophiles. 1998;2(3):297-304. [PubMed ID: 9783177].
-
7.
Ventosa A, Nieto JJ, Oren A. Biology of moderately halophilic aerobic bacteria. Microbiol Mol Biol Rev. 1998;62(2):504-44. [PubMed ID: 9618450].
-
8.
Amoozegar MA, Malekzadeh F, Malik KA. Production of amylase by newly isolated moderate halophile, Halobacillus sp. strain MA-2. Journal of Microbiol Methods. 2003;52(3):353-9. https://doi.org/10.1016/s0167-7012(02)00191-4.
-
9.
Bernhardsdotter ECMJ, Ng JD, Garriott OK, Pusey ML. Enzymic properties of an alkaline chelator-resistant α-amylase from an alkaliphilic Bacillus sp. isolate L1711. Process Biochem. 2005;40(7):2401-8. https://doi.org/10.1016/j.procbio.2004.09.016.
-
10.
Coronado M. Production and biochemical characterization of an α-amylase from the moderate halophile Halomonas meridiana. FEMS Microbiol Lett. 2000;183(1):67-71. https://doi.org/10.1016/s0378-1097(99)00628-x.
-
11.
Kiran KK, Chandra TS. Production of surfactant and detergent-stable, halophilic, and alkalitolerant alpha-amylase by a moderately halophilic Bacillus sp. Strain TSCVKK. Appl Microbiol Biotechnol. 2008;77(5):1023-31. [PubMed ID: 17999060]. https://doi.org/10.1007/s00253-007-1250-z.
-
12.
Sana B, Ghosh D, Saha M, Mukherjee J. Purification and characterization of an extremely dimethylsulfoxide tolerant esterase from a salt-tolerant Bacillus species isolated from the marine environment of the Sundarbans. Process Biochem. 2007;42(12):1571-8. https://doi.org/10.1016/j.procbio.2007.05.026.
-
13.
Torres S, Martinez MA, Pandey A, Castro GR. An organic-solvent-tolerant esterase from thermophilic Bacillus licheniformis S-86. Bioresour Technol. 2009;100(2):896-902. [PubMed ID: 18723341]. https://doi.org/10.1016/j.biortech.2008.07.009.
-
14.
Karbalaei-Heidari HR, Amoozegar MA, Hajighasemi M, Ziaee AA, Ventosa A. Production, optimization and purification of a novel extracellular protease from the moderately halophilic bacterium Halobacillus karajensis. J Ind Microbiol Biotechnol. 2009;36(1):21-7. [PubMed ID: 18781348]. https://doi.org/10.1007/s10295-008-0466-y.
-
15.
Patel RK, Dodia MS, Joshi RH, Singh SP. Purification and characterization of alkaline protease from a newly isolated haloalkaliphilic Bacillus sp. Process Biochem. 2006;41(9):2002-9. https://doi.org/10.1016/j.procbio.2006.04.016.
-
16.
Sanchez-Porro C, Mellado E, Bertoldo C, Antranikian G, Ventosa A. Screening and characterization of the protease CP1 produced by the moderately halophilic bacterium Pseudoalteromonas sp. strain CP76. Extremophiles. 2003;7(3):221-8. [PubMed ID: 12768453]. https://doi.org/10.1007/s00792-003-0316-9.
-
17.
Katoh R, Ngata S, Ozawa A, Ohshima T, Kamekura M, Misono H. Purification and characterization of leucine dehydrogenase from an alkaliphilic halophile, Natronobacterium magadii MS-3. Journal of Molecular Catalysis B: Enzymatic. 2003;23(2-6):231-8. https://doi.org/10.1016/s1381-1177(03)00085-7.
-
18.
Tokunaga H, Arakawa T, Fukada H, Tokunaga M. Opposing effects of NaCl on reversibility and thermal stability of halophilic beta-lactamase from a moderate halophile, Chromohalobacter sp. 560. Biophys Chem. 2006;119(3):316-20. [PubMed ID: 16256261]. https://doi.org/10.1016/j.bpc.2005.10.006.
-
19.
Darvishi P, Ayatollahi S, Mowla D, Niazi A. Biosurfactant production under extreme environmental conditions by an efficient microbial consortium, ERCPPI-2. Colloids Surf B Biointerfaces. 2011;84(2):292-300. [PubMed ID: 21345657]. https://doi.org/10.1016/j.colsurfb.2011.01.011.
-
20.
Wang L, Tang Y, Wang S, Liu RL, Liu MZ, Zhang Y, et al. Isolation and characterization of a novel thermophilic Bacillus strain degrading long-chain n-alkanes. Extremophiles. 2006;10(4):347-56. [PubMed ID: 16604274]. https://doi.org/10.1007/s00792-006-0505-4.
-
21.
Cooper DG, Goldenberg BG. Surface-active agents from two bacillus species. Appl Environ Microbiol. 1987;53(2):224-9. [PubMed ID: 16347271].
-
22.
Folmsbee M, Duncan K, Han SO, Nagle D, Jennings E, McInerney M. Re-identification of the halotolerant, biosurfactant-producing Bacillus licheniformis strain JF-2 as Bacillus mojavensis strain JF-2. Syst Appl Microbiol. 2006;29(8):645-9. [PubMed ID: 16488097]. https://doi.org/10.1016/j.syapm.2006.01.010.
-
23.
George MG. David R B, Richard WC, George MG, Don JB, Noel RK, James TS, editors. Bergey's Manual® of Systematic Bacteriology. Germany: Springer; 2005.
-
24.
Wu XY, Walker MJ, Hornitzky M, Chin J. Development of a group-specific PCR combined with ARDRA for the identification of Bacillus species of environmental significance. J Microbiol Methods. 2006;64(1):107-19. [PubMed ID: 15979744]. https://doi.org/10.1016/j.mimet.2005.04.021.
-
25.
Manachini PL, Mora D, Nicastro G, Parini C, Stackebrandt E, Pukall R, et al. Bacillus thermodenitrifican ssp. nov., nom. rev. IUMS. 2000;50(3):1331-7.
-
26.
Vaerewijck MJ, De Vos P, Lebbe L, Scheldeman P, Hoste B, Heyndrickx M. Occurrence of Bacillus sporothermodurans and other aerobic spore-forming species in feed concentrate for dairy cattle. J Appl Microbiol. 2001;91(6):1074-84. [PubMed ID: 11851816].
-
27.
Kuisiene N, Raugalas J, Citavicius D. Identification of Geobacillus stearothermophilus by restriction digestion with AluI of the amplified 16S rDNA. Biologija. 2007;53(4):62-6.
-
28.
Al-Sulaimani H, Al-Wahaibi Y, Al-Bahry S, Elshafie A, Al-Bemani A, Joshi S, et al. Optimization and Partial Characterization of Biosurfactants Produced by. SPE J. 2011;16(3). https://doi.org/10.2118/129228-pa.
-
29.
Lotfabad TB, Shourian M, Roostaazad R, Najafabadi AR, Adelzadeh MR, Noghabi KA. An efficient biosurfactant-producing bacterium Pseudomonas aeruginosa MR01, isolated from oil excavation areas in south of Iran. Colloids Surf B Biointerfaces. 2009;69(2):183-93. [PubMed ID: 19131218]. https://doi.org/10.1016/j.colsurfb.2008.11.018.
-
30.
Ghojavand H, Vahabzadeh F, Mehranian M, Radmehr M, Shahraki Kh A, Zolfagharian F, et al. Isolation of thermotolerant, halotolerant, facultative biosurfactant-producing bacteria. Appl Microbiol Biotechnol. 2008;80(6):1073-85. [PubMed ID: 18682926]. https://doi.org/10.1007/s00253-008-1570-7.
-
31.
Shavandi M, Mohebali G, Haddadi A, Shakarami H, Nuhi A. Emulsification potential of a newly isolated biosurfactant-producing bacterium, Rhodococcus sp. strain TA6. Colloids Surf B Biointerfaces. 2011;82(2):477-82. [PubMed ID: 21030223]. https://doi.org/10.1016/j.colsurfb.2010.10.005.