Abstract
Background:
Hepatitis B infection, caused by hepatitis B Virus (HBV), is one of the major global public health problems. Hepatitis B Virus genotypes appear to show varying geographic distribution with possible pathogenic and therapeutic differences. Knowledge of HBV genotypes is very important for clinical treatment. Lamivudine is a nucleoside analogue that is clinically used to treat chronic hepatitis B infection. However, the main problem with the application of lamivudine is the development of viral resistance to the treatment with this anti viral drug. Besides, it has been suggested that lamivudine -resistant HBV may be genotype dependent. However, HBV genotype distribution and the biological relevance in this region are poorly understood.Objectives:
The current study aimed to determine hepatitis B genotypes and their correlation with lamivudine- resistant HBV frequency among patients with chronic hepatitis B from Shahrekord, Iran.Methods and Materials:
Hepatitis B virus DNA was detected by conventional PCR in some of the serum samples obtained from HBsAg-positive Chronic Hepatitis B (CHB) patients who were referred to Health Centers of Shahrekord for routine monitoring of the disease. Subsequently, using real-time PCR, the DNA samples were used for genotyping and analysis of resistance to lamivudine.Results:
The DNA was detected in 23 out of 116 (19.82%) of the studied samples. Genotypes D and C were found in 17 out of 23 (73.9%), and in 6 out of 23 (26.1%) of the samples, respectively. To the authors’ best knowledge, the current study is the first report on isolation of Genotype C from Iran. Two out of 17 (11.76%), and 6 out of 6 (100%) of genotypes D and C were resistant to lamivudine, respectively. Resistance to this drug was significantly different between genotypes C and D (P <0.001).Conclusions:
In addition to genotype D, other lamivudine resistant hepatitis B genotypes might be distributed in Iran.Keywords
Hepatitis B Chronic Polymerase Chain Reaction Genotyping Techniques
1. Background
Hepatitis B Virus (HBV) infection is a major global health problem and important infectious disease. It is estimated that there are more than 300 million people worldwide with chronic HBV infection and that 10% of these patients will die as a direct consequence of persistent viral infection (1, 2). The course of infection varies from unapparent self-limiting to chronic active hepatitis which may lead to death after many years (3, 4).
HBV is an error prone DNA virus, which undergoes genetic variability during its replication (5, 6). DNA sequencing of HBV isolates has revealed the existence of 8 viral genotypes A-H (7). HBV genotypes show varying geographic distribution (8). HBV/A is prevalent in Europe, Africa, and India (8, 9). HBV/B and HBV/C are predominant in most parts of Asia, including China and Japan (8, 10).Genotypes B and C are dominant in the Far East and south-east Asia where HBV infection is highly endemic (11, 12).Genotypes A and D are more common in Western Europe and North America. Genotype D is also predominant in the Mediterranean area as well as the Middle East, including India and Iran (13-18).
Response to antiviral treatment may be influenced by HBV genotypes (5-7). Lamivudine, a nucleoside analogue, is widely used for treatment of chronic hepatitis B patients (19-21). However, the major concern is the emergence of lamivudine-resistant HBV during the long term treatment of chronic hepatitis B (22-25). Previous studies indicated that HBV genotypes B and C influenced the response during treatment with interferon-α (26, 27). Similarly, HBV genotypes have played an important role on the response to lamivudine in the treatment of patients with chronic hepatitis B (26, 28).
Based on HBsAg detection, Iran is located in an intermediate endemic region for chronic HBV infection in the Middle East, and patients with chronic HBV infection are presented with different clinical pictures. HBV genotyping studies on HBV-infected individuals from different regions of Iran showed that HBV genotype D is dominant in most regions of Iran (15-18). In Iran, although there are some reports on hepatitis B infection (29-31), there is no data on the distribution of HBV genotypes and genotype-dependent development of resistance to lamivudine in patients with CHB. Therefore, the present study aimed to investigate the distribution of HBV genotypes and its relation with lamivudine resistance in patients with CHB in Shahrekord. The findings of this study would provide epidemiologic data, and possibly a guideline for treatment management of the patients.
2. Objectives
The current study aimed to determine hepatitis B genotypes and their correlation with lamivudine resistance frequency among patients with chronic hepatitis B from Shahrekord, Iran. The present study aimed to determine: 1) The prevalence of HBV genotypes in HBsAg-positive individuals who did not receive lamivudine treatment, 2) the rate of lamivudine resistance in these carriers, and 3) the association of HBV genotype with lamivudine resistance status.
3. Materials and Methods
3.1. Clinical Samples
A total of 116 serum samples were collected from the same number of HBsAg-positive CHB patients who were referred to the health centers of Shahrekord. Thirty six patients were female and 80 were male, with the age ranging from 25 to 71 years. Inclusion criteria were HBsAg-positive patients with anti-HCV and anti-HIV negative markers. Patients were registered irrespective of HBsAg status, ALT level, HBV DNA level or antiviral treatment status.
3.2. PCR Amplification and Detection of HBV DNA
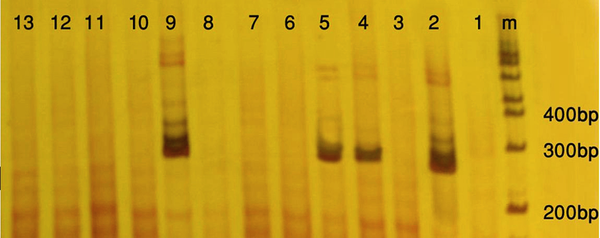
HBV DNA was extracted from the serum samples using an extraction kit (QIAamp MinElute Virus Kits, Qiagen, Germany) and subjected to conventional PCR, using appropriate primers and positive control provided by the kit (plasma-serum HBV PCR detection kit, Norgen-Canada) according to the manufacturer's instructions. Strict measures were adopted to prevent any contamination. An aliquot of water was used as the negative control. Samples were considered positive if they yielded at least two positive results in two different reactions and were considered negative when there were two negative results in two different reactions. To detect the PCR product, a total of 10 microliter of the PCR products was analyzed by electrophoresis through a polyacrylamide gel, stained with silver nitrate and photographed.
3.3. HBV Genotyping and Detection of Lamivudine-Resistant Genotypes
Detection of HBV genotypes and lamivudine resistance was performed using quantitative real time PCR. For these purposes two commercial kits, HBV Genotype B, C and D real-time PCR kit, and HBV YMDD Mutation Real Time PCR Kit (Liferiver, Shanghai, China) were used respectively according to the manufacturer's instructions. All of the applied kits were specific ready-to-use systems that worked based on fluorogenic 5ʹ nuclease assay using specific primers and probes, provided by kits. For real-time PCR a rotor gene 3000 instrument (Corbett Research-Australia) was used. All necessary precautions to prevent cross contamination were observed; also positive and negative controls were included at each step.
For genotyping, briefly, the kit contained two separate master mixes, one for typing of D genotype in FAM channel, and the other one for typing B in FAM channel and C in JOE channel. To detect lamivudine resistance, the kit contained single assay master mixes tube, and specific primers and probes to detect the strains with YMDD wild type motif in both FAM and JOE channels, and also the strains with YIDD or YVDD motif in only JOE channel (but negative on FAM channel). The assay results were compared with those of the kit's positive controls.
4. Results
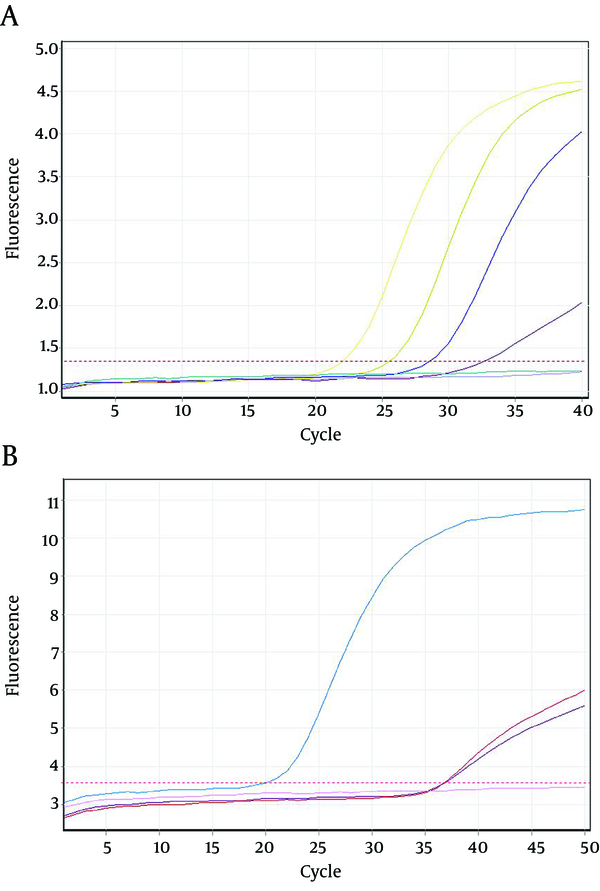
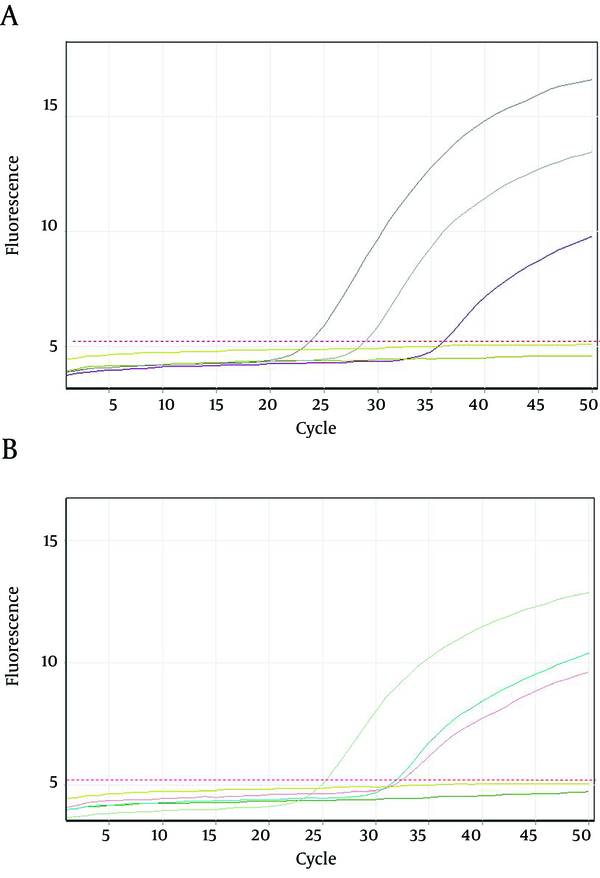
Twenty seven out of 116 (23.2%) of the HBsAg positive individuals were female and their mean age was 39.11 years. Twenty three out of 116 patients (19.82%) were positive for HBV DNA in the conventional PCR (Figure 1). Using real-time PCR, 17 out of 23 (73.9%) were infected with HBV genotype D (Figure 2) and 6 out of 23 (26.1%) were infected with HBV genotype C (Figure 3). To the authors` best knowledge, this is the first report on the isolation of genotype C in Iran. Eight out of 23 (34.7%) of HBV DNA positive samples were resistant to lamivudine. Two out of 17 (11.76%) and 6 out of 6 (100%) of genotypes D and C were resistant to lamivudine respectively. Resistance to this drug was significantly different in genotypes C and D (P < 0.001).
Polyacrylamide Gel Electrophoresis of Hepatitis B Virus PCR Products
Amplification Plots of Hepatitis B Virus D and C Genotype Using Real-time PCR
Amplification Plots of Lamivudine- Resistant Hepatitis B Virus Strains Using Real-Time PCR
5. Discussion
Variation of HBV genome during viral replication leads to creation of different genotypes with different geographical distribution (1, 2). Clinical picture, prognosis of the disease and response to antiviral treatment may be genotype dependent (21). Accordingly, the current study investigated the distribution of HBV genotypes and their resistance to lamivudine in a group of CHB patients who did not receive lamivudine in a central province of Iran.
The current study results showed that genotype D is the most prevalent genotype in this region, since it was found in 73.9% of the CHB patients. Consistent with results of this report, it has been shown that this genotype is the most prevalent genotype worldwide (13, 14) and also in the most regions of Iran (15-18). However, as the current study results showed, it may not be the only genotype distributed in Iran. Based on the results of the current study, 26.1% of these patients were infected with HBV genotype C. To the authors best knowledge, this is the first report on this genotype from Iran. Genotype C is one of the most prevalent genotypes in Thailand and China (8-12). It has been also suggested that distribution of HBV genotype is influenced by immigrant population (8-10). Since a large number of Iranian have traveled to these two countries in the recent years, there is the possibility that distribution of this genotype in our region originated from these two countries.
Error-prone replication of HBV leads to appearance of lamivudine resistant strains of this virus (32, 33). Some published results indicated that lamivudine resistant mutant existed in the serum of CHB patients who did not receive lamivudine treatment (32, 34). In the present study, eight out of 23 (34.7%) of HBV DNA from CHB individuals who did not receive lamivudine, showed this mutation. Therefore, it might be concluded that naturally occurring HBV lamivudine resistant mutants are circulated in the region as well. According to the results of the current study, 11.76% of HBV genotype D and 100% of genotype C were resistant to lamivudine, indicating that lamivudine resistance was significantly different in genotypes C and D (P <0.001). There are some reports indicating that resistant to this antiviral among HBV genotypes is significantly different (24, 26, 28) and that the rate of resistance to this drug is lower in patients infected with genotype D than in patients with some other genotypes which is in agreement with those of the current study findings.
The results of the study might provide epidemiologic data on the distribution of HBV genotypes in this region. In conclusion, based on the obtained results HBV genotype D might not be the only genotype distributed in most regions of Iran, and some other genotypes such as genotype C might be present in this country. In addition, the current study findings, in accordance with many other published ones, would provide supportive evidences indicating that HBV lamivudine resistance is genotype dependent and naturally occurring HBV lamivudine resistant mutants might circulate in our country as well.
Acknowledgements
References
-
1.
Alter MJ. Epidemiology and prevention of hepatitis B. Semin Liver Dis. 2003;23(1):39-46. [PubMed ID: 12616449]. https://doi.org/10.1055/s-2003-37583.
-
2.
Maddrey WC. Hepatitis B--an important public health issue. Clin Lab. 2001;47(1-2):51-5. [PubMed ID: 11214223].
-
3.
Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol. 2004;72(3):363-9. [PubMed ID: 14748059]. https://doi.org/10.1002/jmv.10534.
-
4.
Mahoney FJ. Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin Microbiol Rev. 1999;12(2):351-66. [PubMed ID: 10194463].
-
5.
Hannoun C, Horal P, Lindh M. Long-term mutation rates in the hepatitis B virus genome. J Gen Virol. 2000;81(Pt 1):75-83. [PubMed ID: 10640544].
-
6.
Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83(Pt 6):1267-80. [PubMed ID: 12029141].
-
7.
Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69 ( Pt 10):2575-83. [PubMed ID: 3171552].
-
8.
Liu WC, Phiet PH, Chiang TY, Sun KT, Hung KH, Young KC, et al. Five subgenotypes of hepatitis B virus genotype B with distinct geographic and virological characteristics. Virus Res. 2007;129(2):212-23. [PubMed ID: 17825452]. https://doi.org/10.1016/j.virusres.2007.07.016.
-
9.
Datta S. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. Virol J. 2008;5:156. [PubMed ID: 19099581]. https://doi.org/10.1186/1743-422X-5-156.
-
10.
Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34(3):590-4. [PubMed ID: 11526547]. https://doi.org/10.1053/jhep.2001.27221.
-
11.
Chin R, Locarnini S. Treatment of chronic hepatitis B: current challenges and future directions. Rev Med Virol. 2003;13(4):255-72. [PubMed ID: 12820187]. https://doi.org/10.1002/rmv.393.
-
12.
Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362(9401):2089-94. [PubMed ID: 14697813]. https://doi.org/10.1016/s0140-6736(03)15108-2.
-
13.
Patel PC, Cockcroft DW. Occupational asthma caused by exposure to cooking lobster in the work environment: a case report. Ann Allergy. 1992;68(4):360-1. [PubMed ID: 1558332].
-
14.
Lindh M, Andersson AS, Gusdal A. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus--large-scale analysis using a new genotyping method. J Infect Dis. 1997;175(6):1285-93. [PubMed ID: 9180165].
-
15.
Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Adeli A, Sabahi F, Abachi M, Azizi M, et al. Complete genomic sequence and phylogenetic relatedness of hepatitis B virus isolates from Iran. J Med Virol. 2005;76(3):318-26. [PubMed ID: 15902699]. https://doi.org/10.1002/jmv.20362.
-
16.
Mojiri A, Behzad-Behbahani A, Saberifirozi M, Ardabili M, Beheshti M, Rahsaz M, et al. Hepatitis B virus genotypes in southwest Iran: molecular, serological and clinical outcomes. World J Gastroenterol. 2008;14(10):1510-3. [PubMed ID: 18330939].
-
17.
Bahramali G, Sadeghizadeh M, Amini-Bavil-Olyaee S, Alavian SM, Behzad-Behbahani A, Adeli A, et al. Clinical, virologic and phylogenetic features of hepatitis B infection in Iranian patients. World J Gastroenterol. 2008;14(35):5448-53. [PubMed ID: 18803358].
-
18.
Merat S, Rezvan H, Nouraie M, Jamali A, Assari S, Abolghasemi H, et al. The prevalence of hepatitis B surface antigen and anti-hepatitis B core antibody in Iran: a population-based study. Arch Iran Med. 2009;12(3):225-31. [PubMed ID: 19400598].
-
19.
Severini A, Liu XY, Wilson JS, Tyrrell DL. Mechanism of inhibition of duck hepatitis B virus polymerase by (-)-beta-L-2',3'-dideoxy-3'-thiacytidine. Antimicrob Agents Chemother. 1995;39(7):1430-5. [PubMed ID: 7492080].
-
20.
Dienstag JL, Schiff ER, Wright TL, Perrillo RP, Hann HW, Goodman Z, et al. Lamivudine as initial treatment for chronic hepatitis B in the United States. N Engl J Med. 1999;341(17):1256-63. [PubMed ID: 10528035]. https://doi.org/10.1056/NEJM199910213411702.
-
21.
Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339(2):61-8. [PubMed ID: 9654535]. https://doi.org/10.1056/NEJM199807093390201.
-
22.
Wang JH, Lu SN, Lee CM, Lee JF, Chou YP. Fatal hepatic failure after emergence of the hepatitis B virus mutant during lamivudine therapy in a patient with liver cirrhosis. Scand J Gastroenterol. 2002;37(3):366-9. [PubMed ID: 11916202].
-
23.
Kao JH, Liu CJ, Chen DS. Hepatitis B viral genotypes and lamivudine resistance. J Hepatol. 2002;36(2):303-4. [PubMed ID: 11830346].
-
24.
Papatheodoridis GV, Dimou E, Papadimitropoulos V. Nucleoside analogues for chronic hepatitis B: antiviral efficacy and viral resistance. Am J Gastroenterol. 2002;97(7):1618-28. [PubMed ID: 12135009]. https://doi.org/10.1111/j.1572-0241.2002.05819.x.
-
25.
Bottecchia M, Souto FJ, O KM, Amendola M, Brandao CE, Niel C, et al. Hepatitis B virus genotypes and resistance mutations in patients under long term lamivudine therapy: characterization of genotype G in Brazil. BMC Microbiol. 2008;8:11. [PubMed ID: 18211717]. https://doi.org/10.1186/1471-2180-8-11.
-
26.
Zollner B, Petersen J, Puchhammer-Stockl E, Kletzmayr J, Sterneck M, Fischer L, et al. Viral features of lamivudine resistant hepatitis B genotypes A and D. Hepatology. 2004;39(1):42-50. [PubMed ID: 14752821]. https://doi.org/10.1002/hep.20016.
-
27.
Allen MI, Deslauriers M, Andrews CW, Tipples GA, Walters KA, Tyrrell DL, et al. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Lamivudine Clinical Investigation Group. Hepatology. 1998;27(6):1670-7. [PubMed ID: 9620341]. https://doi.org/10.1002/hep.510270628.
-
28.
Palumbo E. Hepatitis B genotypes and response to antiviral therapy: a review. Am J Ther. 2007;14(3):306-9. [PubMed ID: 17515708]. https://doi.org/10.1097/01.pap.0000249927.67907.eb.
-
29.
Karimi A, Hoseini SM. Seroprevalence of hepatitis B and C virus and HIV markers among blood donors from Shahre-Kord, Iran (2004-2006). Kuwait Med J. 2008;40(4):279-81.
-
30.
Mokhtarian K, Karimi A. Evaluation of Vaccine Induced Immunity to Hepatitis B Virus among Health Care Workers in a University Hospital in Iran. Kuwait Med J. 2010;42:202-4.
-
31.
Karimi A, Imani R. [Prevalence of Hbsag, Hcv And Hiv Antibodies Among Intravenous Drug Users In Shahre-Kord, Iran]. J Isfahan Med School. 2007;8:10-4.
-
32.
Zhang XH, Zhang YX, Sun LR, Wen Q, Zhou LQ, Fan GX, et al. [Study of gene chips in the detection of YMDD mutations in the region of HBV polymeras]. Zhonghua Yi Xue Za Zhi. 2003;83(6):459-62. [PubMed ID: 12887756].
-
33.
Akarsu M, Sengonul A, Tankurt E, Sayiner AA, Topalak O, Akpinar H, et al. YMDD motif variants in inactive hepatitis B carriers detected by Inno-Lipa HBV DR assay. J Gastroenterol Hepatol. 2006;21(12):1783-8. [PubMed ID: 17074014]. https://doi.org/10.1111/j.1440-1746.2006.04567.x.
-
34.
Yan MH, Zhang C, Ling Q, Zhou RF. [Detection of YMDD motif mutations in lamivudine-untreated patients with chronic hepatitis B]. Zhonghua Gan Zang Bing Za Zhi. 2003;11(7):430-1. [PubMed ID: 12890351].