Abstract
Background:
Molecular typing techniques are reliable tools for epidemiological study of tuberculosis because of their power in detecting recent transmission and differentiating reinfection and relapses.Objectives:
The present study investigated epidemiological diversity among Mycobacterium tuberculosis strains circulating in three Khorasan provinces, Iran, using 12-loci MIRU-VNTR and spoligotyping.Methods:
This study was performed on 140 M. tuberculosis strains selected from the sputum of new cases of pulmonary tuberculosis patients in three Khorasan provinces, Iran. 12 loci MIRU-VNTR and Spoligotyping were performed on all isolates.Results:
By MIRU-VNTR analysis, 76 distinct patterns comprising 19 clusters and 57 unique patterns were identified. Based on the results, MIRU10, MIRU26, and ETRF were highly discriminative, ETRD was poorly discriminative and other loci were designated as moderately discriminative. Spoligotyping of isolates revealed 51 distinct patterns: 26 patterns containing 33 strains (23.6%) corresponding to orphan strains and 14 patterns containing 107 strains (76.4%) corresponding to shared-types in the SITVIT2 database. Totally, 103 isolates (73.6%) were classified into 14 clusters containing 2 - 56 isolates; the remaining 37 isolates (26.4%) were unique patterns. By combining two techniques, 94 distinct patterns (15 clusters) contained 61 isolates (43.6%), and 79 unique patterns were identified. The discriminatory power (HGDI) of combination of two techniques was 0.962, which was higher than that of each technique alone. Based on the trees designed by Bionumerics software, we differentiated isolates with similar genetic patterns and grouped them together. Two great clusters were Haarlem and CAS lineage. All strains with combined drug resistance related to Beijing strains. Also, all mono drug-resistant strains related to Haarlem family; other strains were susceptible to the first-line anti-tuberculosis drugs. Also, homoplasy was observed in a number of patterns.Conclusions:
In MIRU-VNTR typing method, according to the genotype of each area, the loci with high discriminatory power (such as miru10, miru26, and ETRF in Iran) are recommended to be used and the loci with poor discrimination (such as ETRD in Iran) are not.Keywords
Genotyping Method VNTR Spoligotyping Mycobacterium tuberculosis
1. Background
Tuberculosis (TB) is now considered as one of the major public health concerns in the world. Also, the emergence of multi-drug resistant (MDR) strains and co-infection with the HIV virus is considered as one of the biggest health problems (1). Globally, 1.5 million people have lost their lives due to this disease and according to the 2015 world health organization report, 9.6 million people are estimated to have fallen ill with TB in 2014. This report also showed that the number of people with TB had increased in the world and emphasized that the number of MDR-TB have increased in the past two decades. From the new TB cases in 2014, 58% were in the Southeast Asia and Western pacific regions (2). As Iran is in the vicinity of TB high-prevalence countries, prevention of the spread of tuberculosis, particularly MDR-TB, is now one of the priorities in the country (3).
For understanding the path of transmission and preventing the spread of the disease molecular epidemiology studies are of great importance (4). In this regard, genotyping techniques are powerful tools to show the outbreak, conduct contact tracing, and study the diversity of strains (5). Nowadays, these typing techniques are reliable tools for epidemiological study of TB because of their power in detecting recent transmission and differentiating re-infections and relapses (6). There are several molecular typing techniques available for studying epidemiology of Mycobacterium tuberculosis (5). Two new typing PCR-based methods [i.e. Mycobacterial interspersed repetitive unit variable number tandem repeat (MIRU-VNTR) and Spacer oligonucleotide typing (spoligotyping)] are the most commonly used techniques (1). MIRU-VNTR typing is a very useful tool for genotyping; among its advantages are high discriminatory power, high reproducibility (7), and the ability to create a numeric code for each isolate which simplifies its tracing in the database as well as ease of use and cost-effectiveness. Also, spoligotyping is a PCR-based method which is based on DNA polymorphism within the direct repeat (DR) locus of M. tuberculosis that allows genotyping M. tuberculosis complex in a fast, reliable, and cost-effective way (8).
In the present study, MIRU-VNTR and spoligotyping were applied for genotyping of M. tuberculosis strains isolated from patients in three Northeast provinces of Iran (i.e. Razavi Khorasan, North Khorasan, and South Khorasan provinces which named the Great Khorasan, the largest province in Iran, before 2000). The three provinces have a total population of 7,524,663 (available at the http://www.amar.org.ir/) that approximately comprise ten percent of the Iranian population. Several studies have been performed on the subject in Iran although information on the genetic diversity of M. tuberculosis in this region is rare.
2. Objectives
This study, therefore, aimed at genotyping and tracking the transmission dynamics of M. tuberculosis isolates from three Northeast provinces of Iran (i.e. Razavi Khorasan, North Khorasan, and South Khorasan provinces), using 12-locus MIRU-VNTR and spoligotyping.
3. Methods
3.1. Bacterial Isolates and Clinical Data
This cross-sectional study was carried out on 140 M. tuberculosis positive cultures obtained from new cases of pulmonary TB in three Khorasan provinces of Iran from April to September 2014. Sputum samples were collected from all individuals residing in the urban and rural areas of the three Khorasan provinces with no history of TB test, with suspicious TB symptoms, or with TB history in one of their family members. Then, positive cases were referred to a reference laboratory in the Northeastern Iran following microscopic sputum examination. In sum, 180 positive cases were collected; however, 40 cases did not meet the inclusion criteria for being NTM or negative culture results.
Demographic information such as age, sex, place of birth, previous TB history, and relevant medical data were collected by the staff of Tuberculosis Center, and then recorded in corresponding forms. Ethical approval for the study was obtained from the ethics committee of Mashhad University of medical sciences (project ethics code 930504). We performed this study on the Mycobacterial isolates that had been collected for another project evaluating antibiotics susceptibility of Mycobacteria in this region.
Primary isolation and culture from sputum samples were performed in accordance with standard protocols (9). Initially, biochemical methods were used to identify isolates (10). Drug susceptibility testing against Rifampin, Isoniazid, and Ethambutol was performed based on the standard method of CLSI (11). Genomic DNA of all positive cultures was extracted as described previously (12-14). They were obtained from suspended colonies in 200 μL of TB lysis® Solution (SinaClon BioScience Company, Iran) by heating at 96°C for 30 minutes and, after centrifugation of the suspension at 9750 g for 15 minutes, the supernatant containing the DNA was removed and stored at -20°C for later use.
3.2. MIRU-VNTR
Twelve loci MIRU-VNTR was performed by using a combination of existing proposed protocols (15, 16). The 12 studied loci were ETR-A, ETR-B, ETR-C, ETR-D, ETR-E, ETR-F, MIRU10, MIRU16, MIRU26, MIRU39, MIRU40, and QUB11b. The sequences of MIRU-VNTR primers used in this study were obtained from Macrogen Company, South Korea.
The PCR conditions for QUB11b locus were 30 cycles of 1 minute at 94°C, 1 minute at 65°C, and 1 minute at 72°C, and for other loci were 40 cycles of 1 minute at 94°C, 1 minute at 68°C and, 1 minute at 72°C. The first denaturation and final extension steps for all loci were performed for 10 minute. In order to perform PCR tests, Amplicon commercial kit (Amplicon, Denmark), which contained a single polymerase enzyme, nucleotides Quartet, magnesium chloride and other common salts, was used. PCR was performed in 12 μL volume containing 2 μL DNA template, 6 μL PCR master mix, 0.6 μL specific primers, 0.4 μL DMSO, and 3 μL PCR water.
3.3. Spoligotyping
Spoligotyping was performed for all isolates using the standard method as previously described by Kamerbeek et al. (17). In brief, Direct Repeat (DR) region was amplified by PCR using primers DRa (5’-CCG AGA GGG GAC GGA AAC- 3’) (Biotinylated 5’ end) and DRb (5’- GGT TTT GGG TCT GAC GAC-3’) derived from DR sequence (Isogen Bioscience BV, Maarssen, The Netherlands) and PCR Master Mix (Amplicon, Denmark). PCR products were hybridized to a set of 43 immobilized oligonucleotides, which were derived from the spacer sequences of M. tuberculosis H37RV and M. bovis BCG P3 by reverse line blotting. Hybridized DNA was detected by ECL (Amersham, UK) as well as by exposing the membrane to ECL-Hyper film for 1 - 2 minutes. For positive controls, DNA extracts of M. tuberculosis H37Rv and M. bovis BCG were used. The profiles obtained were analyzed and compared to the world spoligotyping database (18, 19).
3.4. Molecular Epidemiology Analysis
The allelic diversity and genetic relationships among the isolates of each VNTR locus and spoligo patterns were estimated using Bionumerics software. Cluster-graph and minimum spanning tree (MST), which is based on a spoligotyping and VNTR combined distance matrix, were used to analyze the clonal genotypes of studied strains (20).
3.5. Statistical Analyses
The obtained data were analyzed using the SPSS software. Comparison of categorical variables in different groups was performed using the Pearson, Chi-square test, and two-tailed Fisher’s exact test. P-values less than 0.05 were considered to be statistically significant.
4. Results
4.1. Study Population
The study involved 140 M. tuberculosis positive cultures, obtained from new cases of pulmonary TB in three Khorasan provinces of Iran during a six month period from April 2014 to September 2014. From these, 128 (91.4%) belonged to Iranians and 12 (8.6%) to immigrant patients; also, 84 (60%) were from female and 56 (40%) from male patients. The mean age was 53.43 (± 23.37) and 50.65 (± 20.01) years for female and male TB patients, respectively.
The results of drug-susceptibility testing showed that 134 samples (95.7%) were susceptible, 2 samples (1.43%) were MDR-TB cases (INH + RF + ETB resistance), three samples (2.14%) had mono drug resistant strains (including two INH and one RF), and one (0.7%) had combined INH and RF resistance.
4.2. MIRU-VNTR Typing
By MIRU-VNTR analysis of the 140 isolates, 76 distinct patterns comprising 19 clusters (10 clusters of 2 strains each, 5 clusters of 3 strains each, 2 clusters of 4 strains, 1 cluster of 13 strains, and 1 cluster of 27 strains) and 57 unique patterns were identified (Tables 1 and 2).
MIRU-VNTR Patterns of Strains from Northeast of Iran at Different Loci (Clustered Isolates)
| No. of Strains | ETR A | ETR B | ETR C | ETR D | ETR E | ETR F | QUB11b | MIRU 10 | MIRU 16 | MIRU 26 | MIRU 39 | MIRU 40 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 6 | 2 | 3 |
| 13 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 3 |
| 4 | 2 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 3 |
| 3 | 3 | 1 | 4 | 3 | 3 | 3.2 | 4 | 3 | 3.2 | 5 | 2 | 1 |
| 3 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 6 | 2 | 2 |
| 3 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 1 |
| 3 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 4 | 2 | 4 |
| 3 | 3 | 1 | 4 | 3 | 3 | 1.2 | 2 | 2 | 3.2 | 5 | 2 | 3 |
| 3 | 2 | 2 | 2 | 3 | 5 | 3.2 | 2 | 5 | 5.2 | 7 | 5 | 3 |
| 2 | 4 | 2 | 5 | 3 | 5 | 3.2 | 6 | 3 | 3.2 | 5 | 3 | 3 |
| 2 | 4 | 2 | 4 | 3 | 5 | 3.1 | 6 | 3 | 3.2 | 5 | 3 | 3 |
| 2 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 6 | 4.2 | 7 | 3 | 3 |
| 2 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 5 | 5.2 | 8 | 3 | 3 |
| 2 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 5 | 4.2 | 7 | 2 | 3 |
| 2 | 4 | 2 | 2 | 3 | 4 | 3.2 | 2 | 5 | 4.2 | 7 | 3 | 4 |
| 2 | 3 | 1 | 4 | 3 | 3 | 2.2 | 3 | 2 | 3.2 | 5 | 2 | 3 |
| 2 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 3 | 3.2 | 6 | 2 | 3 |
| 2 | 3 | 1 | 4 | 3 | 3 | 2.1 | 2 | 2 | 3.2 | 5 | 2 | 3 |
| 2 | 3 | 1 | 4 | 3 | 3 | 2.1 | 1 | 2 | 3.2 | 5 | 2 | 3 |
MIRU-VNTR Patterns of Strains from Northeast of Iran at Different Loci (Non-Clustered Isolates)
| No. of Strains | ETR A | ETR B | ETR C | ETR D | ETR E | ETR F | QUB11b | MIRU 10 | MIRU 16 | MIRU 26 | MIRU 39 | MIRU 40 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | two bound | 1 | 2 | 3 | 5 | 2.2 | 2 | 2 | 4.2 | 5 | 2 | 2 |
| 1 | 4 | 2 | 4 | 3 | 6 | 3.2 | 6 | 3 | 3.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 4 | 3 | 6 | 2.3 | 6 | 3 | 3.2 | 5 | 3 | 0 |
| 1 | 4 | 2 | 4 | 3 | 5 | 3.2 | 8 | 3 | 3.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 4 | 3 | 5 | 3.2 | 6 | 3 | 2.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 4 | 3 | 5 | 2.3 | 6 | 3 | 3.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 2 | 4 | 4 | 3.2 | 2 | 6 | 3.2 | 9 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 6 | 3.2 | 2 | 4 | 4.2 | 4 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.3 | 2 | 7 | 4.2 | 7 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 6 | 4.2 | 5 | 3 | 4 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 6 | 4.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 6 | 4.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 5 | 4.2 | 6 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 2 | 3 | 4.2 | 7 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 5 | 3.2 | 1 | 6 | 3.2 | 7 | 3 | 2 |
| 1 | 4 | 2 | 2 | 3 | 5 | 1.5 | 2 | 6 | 4.2 | 5 | 3 | 4 |
| 1 | 4 | 2 | 2 | 3 | 5 | 1.5 | 2 | 6 | 4.2 | 2 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 4 | 3.2 | 2 | 6 | 4.2 | 7 | 2 | 3 |
| 1 | 4 | 2 | 2 | 3 | 4 | 3.2 | 2 | 5 | 3.2 | 8 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 4 | 3.1 | 2 | 5 | 4.2 | 7 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 4 | 1.5 | 2 | 5 | 5.2 | 9 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 3 | 3.2 | 2 | 5 | 6.2 | 5 | 3 | 3 |
| 1 | 4 | 2 | 2 | 3 | 2 | 3.2 | 2 | 6 | 3.2 | 7 | 3 | 0 |
| 1 | 4 | 2 | 2 | 2 | 5 | 1.5 | 2 | 5 | 5.2 | 8 | 3 | 3 |
| 1 | 4 | 1 | 2 | 3 | 5 | 3.2 | 2 | 2 | 4.2 | 7 | 3 | 3 |
| 1 | 3 | 2 | 4 | 3 | 4 | 1.3 | 3 | 2 | 1.2 | 6 | 2 | 0 |
| 1 | 3 | 2 | 4 | 3 | 3 | 3.2 | 5 | 3 | 3.2 | 5 | 2 | 4 |
| 1 | 3 | 2 | 4 | 3 | 3 | 1.5 | 3 | 3 | 1.2 | 5 | 2 | 3 |
| 1 | 3 | 2 | 4 | 3 | 3 | 1.3 | 3 | 3 | 1.2 | 5 | 2 | 5 |
| 1 | 3 | 2 | 4 | 2 | 3 | 2.1 | 2 | 5 | 1.2 | 1 | 2 | 2 |
| 1 | 3 | 2 | 3 | 3 | 3 | 3.2 | 7 | 4 | 3.2 | 5 | 2 | 3 |
| 1 | 3 | 2 | 3 | 3 | 3 | 3.2 | 6 | 5 | 3.2 | 5 | 2 | 3 |
| 1 | 3 | 2 | 3 | 3 | 3 | 3.2 | 3 | 3 | 3.2 | 5 | 2 | 3 |
| 1 | 3 | 2 | 3 | 3 | 3 | 1.5 | 7 | 4 | 3.2 | 5 | 2 | 3 |
| 1 | 3 | 2 | 2 | 3 | 4 | 3.2 | 2 | 6 | Three bound | 5 | 3 | 3 |
| 1 | 3 | 1 | 6 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 1 |
| 1 | 3 | 1 | 5 | 3 | 3 | 2.2 | 1 | 2 | 4.2 | 6 | 2 | 3 |
| 1 | 3 | 1 | 4 | 5 | 3 | 2.2 | 2 | 2 | 3.2 | 6 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 5 | 2.2 | 2 | 2 | 2.2 | 5 | 2 | 2 |
| 1 | 3 | 1 | 4 | 3 | 4 | 2.2 | 2 | 2 | 3.2 | 6 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 4 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.2 | 3 | 2 | 3.2 | 5 | 2 | 2 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 3 | 3.2 | 3 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 4.2 | 6 | 2 | 4 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 5 | 2 | 4 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 3 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.1 | 2 | 2 | 3.2 | 6 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.1 | 2 | 2 | 2.2 | 5 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 2.1 | 2 | 2 | 1.2 | 6 | 2 | 3 |
| 1 | 3 | 1 | 4 | 3 | 3 | 1.1 | 1 | 2 | 3.2 | 5 | 2 | 2 |
| 1 | 2 | 2 | 4 | 3 | 3 | 3.2 | 2 | 3 | 1.2 | 5 | 2 | 4 |
| 1 | 2 | 2 | 4 | 3 | 3 | 3.1 | 2 | 4 | 1.2 | 1 | 2 | 2 |
| 1 | 2 | 2 | 2 | 3 | 6 | 3.2 | 2 | 5 | 4.2 | 7 | 5 | 3 |
| 1 | 2 | 1 | 4 | 3 | 3 | 2.2 | 2 | 2 | 3.2 | 13 | 2 | 3 |
| 1 | 2 | 1 | 4 | 3 | 3 | 2.1 | 2 | 2 | 2.2 | 5 | 2 | 3 |
| 1 | 1 | 2 | 3 | 4 | 3 | 3.2 | 2 | 2 | 3.2 | 4 | 2 | 2 |
| 1 | 0 | 2 | 4 | 3 | 6 | 3.2 | 0 | 6 | 4.2 | 5 | 3 | 3 |
| 1 | 0 | 2 | 2 | 3 | 5 | 3.2 | 0 | 5 | 4.2 | 7 | 3 | 3 |
Based on the results, the allelic diversities of MIRU10, MIRU26, and ETRF were highly discriminative [h-index more than or equal to 0.6 (≥ 0.6)], ETRD was poorly discriminative [h-index less than 0.3 (< 0.3)], and other loci were designated as moderately discriminative [h-index more than 0.3 and less than 0.6 (0.3 < h-index < 0.6)]. The discriminatory power of MIRU-VNTR typing was high (HGDI: 0.951) for these samples.
4.3. Spoligotyping
Spoligotyping of the 140 isolates revealed a total of 51 distinct patterns composed of 26 patterns [containing 33 strains (23.6%)] corresponding to orphan strains in the SITVIT2 database, not yet reported, and 14 patterns [containing 107 strains (76.4%)] corresponding to shared-types or SITs in the SITVIT2 database. In total, 103 isolates (73.6%) were classified into 14 clusters containing 2 - 56 isolates and the remaining 37 isolates (26.4%) were unique patterns. The major observed spoligotype families were ranked as follows: Haarlem, 67/140 (47.9%); Central Asian strain (CAS), 19/140 (13.6%); Beijing, 9/140 (6.4%); U, 5/140 (3.6%), and T, 3/140 (2.1%). The description of the 51 different patterns is shown in Figures 1 and 2.
Description of 25 Shared Types and Corresponding Spoligotyping-Defined Lineages/Sub-Lineages Starting from a Total of 140 Mycobacterium tuberculosis Clinical Isolates from Northeast of Iran
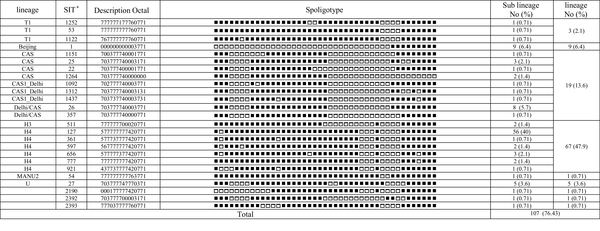
Description of 25 Orphan Types from a Total of 140 Mycobacterium tuberculosis Clinical Isolates from Northeast of Iran
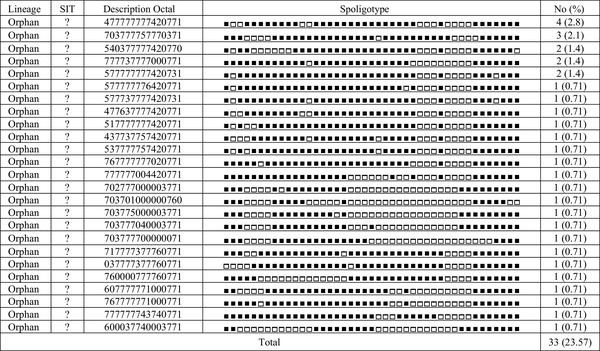
4.4. Molecular Epidemiology Relationships
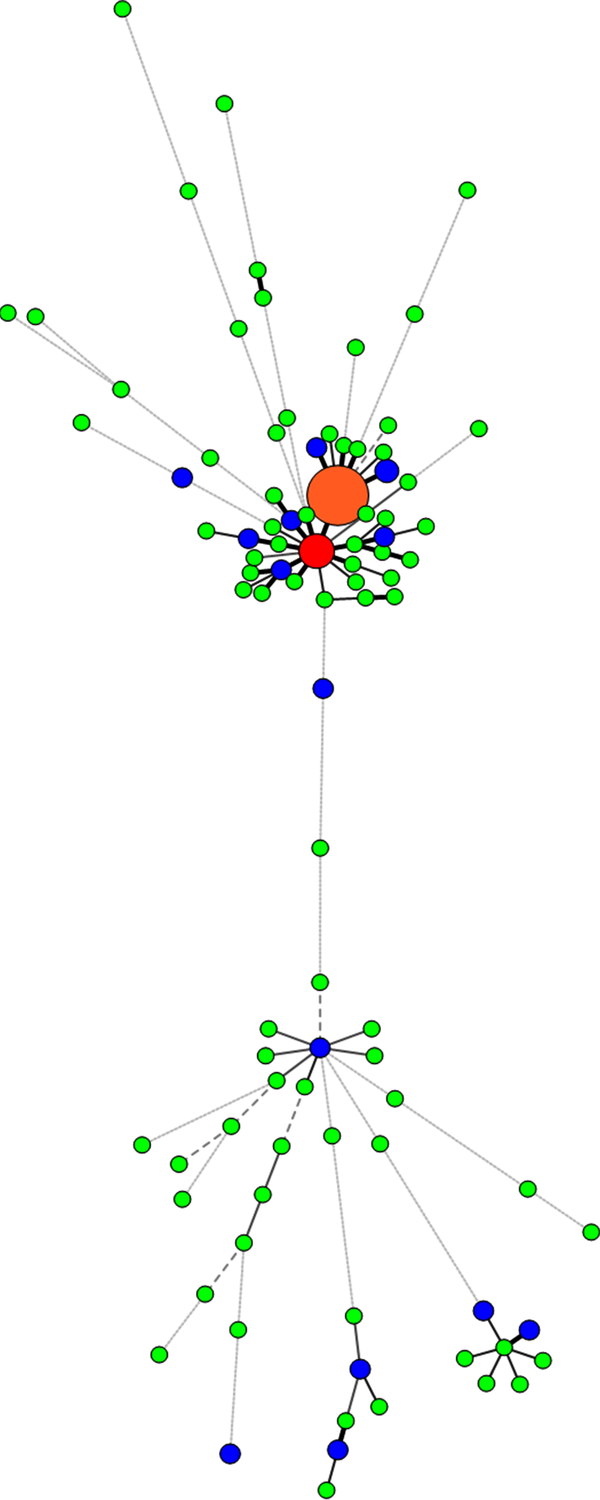
With the help of trees designed by the Bionumerics software, we were able to differentiate isolates with similar genetic patterns and group them together. Two of our very close clusters were Haarlem and CAS lineages; the Haarlem lineage strains constitute the biggest group of strains infecting patients in the great Khorasan province; the major shared types were SIT127, 656, 511, 777, 361, and 921. To confirm lineage categorization, a minimum spanning tree (MST) was derived (Figures 3 and 4).
Spoligotyping and MIRU-VNTR UPGMA Phylogeny Tree

MIRU-VNTR and Spoligotyping Minimum Spanning Tree
5. Discussion
The great Khorasan province, made up of three provinces, is located in the neighborhood of Afghanistan and Pakistan, two of countries that are among 22 TB high-burden countries; according to WHO report (2015), the TB prevalence rate in Afghanistan and Pakistan was 380 per 100,000 population in 2014 (2). Also, high rates of immigration to this region from other parts of Iran as well as its neighboring countries such as Afghanistan are reported. However, despite the high prevalence of TB, there is limited information on the strains prevalent in this area. In the present study, the 140 M. tuberculosis isolates from Northeast of Iran were identified by using Spoligotyping and MIRU-VNTR methods.
The molecular typing of strains by Spoligotyping could not act as a specific indicator for other members of M. tuberculosis complex. The Haarlem family was recognized as the most common type, and the major shared types of Haarlem family were SIT127. It was followed by the CAS family that was ranked as the second most prevalent Spoligotype. Previously published studies from Iran also reported that Haarlem and CAS were the most predominant families (6, 21). Also, in another study performed in Iran, it was shown that Haarlem was the most frequent lineage but the second most prevalent Spoligotype was reported the Beijing genotype (23.5%), followed by the CAS family (22). This difference may be due to the used strains, which were obtained from patients with MDR-TB and were highly associated with the Beijing genotype. Nine of the 140 studied strains (6.4%) in the present study were the Beijing genotype. This genotype was the third most predominant family in the present study. Also, other studies from Iran showed nearly similar corresponding results, e.g. 6.3% in Ramazanzadeh and colleagues (23), 5.3% in Merza et al. (24), and 8.1% in Mozafari et al. (8). The study of Mohajeri et al. (25) showed that the rate of Beijing genotype in the western provinces of Iran was 10.4%. It seems that the frequency of the Beijing genotype in the western provinces of Iran is higher than other areas of the country.
In the present study, 84 out of 140 cases (60%) were female, and 56 (40%) were male. In the Haarlem family, 39.4% were male and 60.6% were female. CAS family contained 31.6% male and 68.4% female. Unlike other lineages, in the Beijing family, 66.7% were male and 33.3% were female. (Male to female ratio was 2 to 1). But there was no significant relationship between sex and lineage (Chi-square: 5.656, P value: 0.463). Study on Afghan refugees in Iran showed that the predominant genotypes were Haarlem and CAS families (6). Since Khorasan provinces are located in the Northeast of the country, and share geographic borders with Afghanistan, these family strains may have been transported from Afghanistan to Khorasan, Iran.
The genotyping pattern obtained from the South Khorasan province (Haarlem 14.3%, CAS 42.9%) was different from the North and Razavi Khorasan provinces (Haarlem 48.9%, CAS 12.1%). It was, however, similar to the pattern obtained from Sistan and Baluchistan province (Southeast of Iran) (20) and Pakistan (26). The South Khorasan province is in the vicinity of Sistan and Baluchistan province and Pakistan country. It seems that these family strains have been most probably transported from these two regions to South Khorasan. In the present study, there was a significant relationship between city of residence and Beijing and CAS lineages (P value: 0.040 for Beijing lineage and 0.032 for CAS lineage). However, there was no significant relationship between city of residence and other lineages. The same 140 isolates were analyzed using the MIRU-VNTR technique and 77 different profiles were observed. 59.3% (83/140) of the isolates were distributed into 19 clusters with 100% of identity, containing 10 clusters of 2 strains each, 5 clusters of 3 strains each, 2 clusters of 4 strains, 1 cluster of 13 strains, and 1 cluster of 27 strains. Based on this study, MIRU10, MIRU26, and ETRF were highly discriminating, but ETRD was poorly discriminating. Most of the previous studies in Iran similarly found that MIRU10, MIRU26, and ETRF were highly discriminating, and MIRU4 or ETRD was poorly discriminating (1, 27-30). By combining the two techniques, 94 distinct patterns comprising 15 clusters containing 61 isolates (43.6%), and 79 unique patterns were identified.
In 10 clusters, some strains exhibited exactly the same profile typing for two methods and were classified as real clusters. Some strains (20 isolates) in the other 9 MIRU-VNTR clusters, however, exhibited different Spoligotype profiles. These different types are named homoplasy. Interestingly, one of the cases of homoplasy was about the two samples from two patients, one of them isolated from the youngest patient of the study (15 years old), and the other one from the oldest patient (91 years old), both living in Mashhad, the center of Razavi Khorasan province. The strains of both of them had the same MIRU-VNTR profile but different types of Spoligotype profiles. Because the half-life of MIRU-VNTR profile is shorter than that of Spoligotyping profile (31), different observed MIRU-VNTR profiles are more reliable and natural in comparison with Spoligotyping patterns. Similar MIRU-VNTR profiles with different Spoligotype profiles are named homoplasy. In this phenomenon, an independent mutational event in a particular spacer leads to the loss of the spacer and makes an invalid Spoligotype pattern (32).
All strains with MDR and combined (INH + RIF) resistance, in the present study, were related to the Beijing strains, confirming its tendency to resist antibiotics. Also, all mono drug resistant strains were relevant to the Haarlem family, but other strains were susceptible to the first-line anti-TB drugs. However, there was no significant relationship between drug resistance and lineage (Fisher’s Exact Test: 218.725, P value: 0.133). Also, the relationship between age and lineage was not significant (Fisher’s Exact Test: 13.040, P value: 0.588). In addition, the discriminatory power (HGDI) of Spoligotyping method was 0.832, the discriminatory power (HGDI) of MIRU-VNTR method was 0.951, and the discriminatory power (HGDI) of the combined two techniques was 0.962, which was higher than that of each technique alone. Therefore, the results showed that in molecular typing of M. tuberculosis, the simultaneous use of the two methods of “Spoligotyping” and “MIRU-VNTR” could increase the genetic patterns of M.tuberculosis strains much effectively compared to the use of only one method.
5.1. Conclusions
MIRU-VNTR is a reproducible method, potentially applicable in tracking epidemiological events such as transmission or relapse. It also allows direct comparison of results between laboratories. In MIRU-VNTR typing method, according to the genotype of each area, the loci with high discriminatory power (such as miru10, miru26, and ETRF in Iran) are recommended to be used and the loci with poor discrimination (such as ETRD in Iran) are not recommended. This typing technique is also accepted as a first line method for molecular epidemiology of M. tuberculosis.
To reduce the amount of homoplasy with these methods, it is recommended to use sequencing-based methods simultaneously.
Acknowledgements
References
-
1.
Zamani S, Aflaki M, Fooladi AA, Darban-Sarokhalil D, Bameri Z, Khazaee S, et al. MIRU-VNTR analysis of the Mycobacterium tuberculosis isolates from three provinces of Iran. Scand J Infect Dis. 2013;45(2):124-30. [PubMed ID: 22954102]. https://doi.org/10.3109/00365548.2012.717233.
-
2.
Global tuberculosis report 2015. Geneva: World Health Organization; 2015.
-
3.
Nasiri MJ, Dabiri H, Darban-Sarokhalil D, Rezadehbashi M, Zamani S. Prevalence of drug-resistant tuberculosis in Iran: systematic review and meta-analysis. Am J Infect Control. 2014;42(11):1212-8. [PubMed ID: 25242634]. https://doi.org/10.1016/j.ajic.2014.07.017.
-
4.
Asgharzadeh M, Kafil H, Pourostadi M. Source case identification and control of tuberculosis by molecular epidemiology, J. Mazandaran Univ Med Sci. 2014;24(115):180-91.
-
5.
Mostrom P, Gordon M, Sola C, Ridell M, Rastogi N. Methods used in the molecular epidemiology of tuberculosis. Clin Microbiol Infect. 2002;8(11):694-704. [PubMed ID: 12445006].
-
6.
Torkaman MR, Nasiri MJ, Farnia P, Shahhosseiny MH, Mozafari M, Velayati AA. Estimation of Recent Transmission of Mycobacterium Tuberculosis Strains among Iranian and Afghan Immigrants: A Cluster-Based Study. J Clin Diagn Res. 2014;8(9):DC05-8. [PubMed ID: 25386431]. https://doi.org/10.7860/JCDR/2014/8886.4864.
-
7.
Kremer K, Arnold C, Cataldi A, Gutierrez MC, Haas WH, Panaiotov S, et al. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J Clin Microbiol. 2005;43(11):5628-38. [PubMed ID: 16272496]. https://doi.org/10.1128/JCM.43.11.5628-5638.2005.
-
8.
Mozafari M, Farnia P, Afraei M, Derakhshani-Nezhad Z, Masjedi MR, Velayati AA. Molecular diversity of Mycobacterium tuberculosis strains indifferent provinces of Iran. Iran J Microbiol. 2013;5(4):366-73. [PubMed ID: 25848506].
-
9.
Githui WA, Meme HK, Juma ES, Kinyanjui P, Karimi F, Chakaya JM, et al. Isolation of multidrug-resistant tuberculosis strains in patients from private and public health care facilities in Nairobi, Kenya. Int J Tuberc Lung Dis. 2004;8(7):837-41. [PubMed ID: 15260274].
-
10.
Farnia P, Masjedi MR, Varahram M, Mirsaeidi M, Ahmadi M, Khazampour M, et al. The recent-transmission of Mycobacterium tuberculosis strains among Iranian and Afghan relapse cases: a DNA-fingerprinting using RFLP and spoligotyping. BMC Infect Dis. 2008;8:109. [PubMed ID: 18681980]. https://doi.org/10.1186/1471-2334-8-109.
-
11.
Rieder HL, Chonde TM, Myking H, Urbanczik R, Laszlo A, Kim SJ, et al. The public health service national tuberculosis reference laboratory and the national laboratory network; minimum requirements, role and operation in a low-income country. IUATLD. 1998:674.
-
12.
Kox LF, Rhienthong D, Miranda AM, Udomsantisuk N, Ellis K, van Leeuwen J, et al. A more reliable PCR for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1994;32(3):672-8. [PubMed ID: 8195377].
-
13.
Kolk AH, Schuitema AR, Kuijper S, van Leeuwen J, Hermans PW, van Embden JD, et al. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbiol. 1992;30(10):2567-75. [PubMed ID: 1400955].
-
14.
Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, et al. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect. 2014;3(3). e19. [PubMed ID: 26038513]. https://doi.org/10.1038/emi.2014.21.
-
15.
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44(12):4498-510. [PubMed ID: 17005759]. https://doi.org/10.1128/JCM.01392-06.
-
16.
Sola C, Filliol I, Legrand E, Lesjean S, Locht C, Supply P, et al. Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics. Infect Genet Evol. 2003;3(2):125-33. [PubMed ID: 12809807].
-
17.
Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907-14. [PubMed ID: 9157152].
-
18.
Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, et al. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int J Tuberc Lung Dis. 2001;5(3):216-9. [PubMed ID: 11326819].
-
19.
Sola C, Filliol I, Gutierrez MC, Mokrousov I, Vincent V, Rastogi N. Spoligotype database of Mycobacterium tuberculosis: biogeographic distribution of shared types and epidemiologic and phylogenetic perspectives. Emerg Infect Dis. 2001;7(3):390-6. [PubMed ID: 11384514]. https://doi.org/10.3201/eid0703.010304.
-
20.
Guernier V, Sola C, Brudey K, Guegan JF, Rastogi N. Use of cluster-graphs from spoligotyping data to study genotype similarities and a comparison of three indices to quantify recent tuberculosis transmission among culture positive cases in French Guiana during a eight year period. BMC Infect Dis. 2008;8:46. [PubMed ID: 18410681]. https://doi.org/10.1186/1471-2334-8-46.
-
21.
Haeili M, Darban-Sarokhalil D, Fooladi AA, Javadpour S, Hashemi A, Siavoshi F, et al. Spoligotyping and drug resistance patterns of Mycobacterium tuberculosis isolates from five provinces of Iran. Microbiologyopen. 2013;2(6):988-96. [PubMed ID: 24311556]. https://doi.org/10.1002/mbo3.139.
-
22.
Farnia P, Masjedi MR, Mirsaeidi M, Mohammadi F, Jallaledin G, Vincent V, et al. Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patients. J Infect. 2006;53(5):331-6. [PubMed ID: 16476483]. https://doi.org/10.1016/j.jinf.2005.12.020.
-
23.
Ramazanzadeh R, Farnia P, Amirmozafari N. Characterization of Mycobacterium tuberculosis complex isolated from iranian and afghani patients by spoligotyping method. Braz J Microbiol. 2009;40(2):314-20. [PubMed ID: 24031364]. https://doi.org/10.1590/S1517-838220090002000019.
-
24.
Merza MA, Farnia P, Salih AM, Masjedi MR, Velayati AA. The most predominant spoligopatterns of Mycobacterium tuberculosis isolates among Iranian, Afghan-immigrant, Pakistani and Turkish tuberculosis patients: a comparative analysis. Chemotherapy. 2010;56(3):248-57. [PubMed ID: 20551642]. https://doi.org/10.1159/000316846.
-
25.
Mohajeri P, Moradi S, Atashi S, Farahani A. Mycobacterium tuberculosis Beijing Genotype in Western Iran: Distribution and Drug Resistance. JCDR. 2016;10(10):5-7.
-
26.
Hasan Z, Tanveer M, Kanji A, Hasan Q, Ghebremichael S, Hasan R. Spoligotyping of Mycobacterium tuberculosis isolates from Pakistan reveals predominance of Central Asian Strain 1 and Beijing isolates. J Clin Microbiol. 2006;44(5):1763-8. [PubMed ID: 16672404]. https://doi.org/10.1128/JCM.44.5.1763-1768.2006.
-
27.
Asgharzadeh M, Kafil HS, Roudsary AA, Hanifi GR. Tuberculosis transmission in Northwest of Iran: using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect Genet Evol. 2011;11(1):124-31. [PubMed ID: 20951237]. https://doi.org/10.1016/j.meegid.2010.09.013.
-
28.
Vatani S, Khosravi AD, Feizabadi MM, Jolodar A. Study of genetic diversity in Mycobacterium tuberculosis by using mycobacterial interspersed repetitive unit: Variable number tandem repeat typing in Khuzestan Province, Iran. Afr J Microbiol Res. 2011;5(12):1549-56.
-
29.
Jafarian M, Aghali-Merza M, Farnia P, Ahmadi M, Masjedi MR, Velayati AA. Synchronous Comparison of Mycobacterium tuberculosis Epidemiology Strains by "MIRU-VNTR" and "MIRU-VNTR and Spoligotyping" Technique. Avicenna J Med Biotechnol. 2010;2(3):145-52. [PubMed ID: 23408229].
-
30.
Ahmadi M, Tadayon K, Mosavari N, Farazi AA, Arjomandzadegan M, Keshavarz R, et al. Mycobacterium tuberculosis genotyping by MIRU-VNTR method. J Gorgan Univ Med Sci. 2015;17(1):Pe97-Pe106. En107.
-
31.
Jagielski T, van Ingen J, Rastogi N, Dziadek J, Mazur PK, Bielecki J. Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. Biomed Res Int. 2014;2014:645802. [PubMed ID: 24527454]. https://doi.org/10.1155/2014/645802.
-
32.
Kato-Maeda M, Gagneux S, Flores LL, Kim EY, Small PM, Desmond EP, et al. Strain classification of Mycobacterium tuberculosis: congruence between large sequence polymorphisms and spoligotypes. Int J Tuberc Lung Dis. 2011;15(1):131-3. [PubMed ID: 21276309].