Abstract
Background:
There should be a public environmental reservoir for Helicobacter pylori in the developing countries, such as Iran, due to their high infection rate of over 70%. Epidemiological findings revealed that water could be a possible source of H. pylori transmission. However, high prevalence of H. pylori in drinking water in Kermanshah, West of Iran, was detected in the authors’ previously published study. The current study aims at designing a more accurate and rapid procedure to investigate the prevalence of Helicobacter species and cagA gene in drinking water samples in Kermanshah, from October to December 2012.Methods:
In the current study, 60 tap water samples were obtained and specific polymerase chain reaction (PCR) targeted cagA and 16s rRNA was performed. A loop-mediated isothermal amplification (LAMP) targeted ureC gene was developed to accurately detect H. pylori in water samples.Results:
The prevalence of ureC by PCR, ureC by LAMP and 16s rRNA by PCR were 26.67%, 38%, and 61.67%, respectively. Among 24 samples (40%), 1 of the 2 tests was positive. The prevalence of cagA gene among ureC positive, 16s rRNA positive and all samples were 18.75%, 13.51%, and 10%, respectively.Conclusions:
Helicobacter pylori contamination in drinking water was considerably higher using LAMP compared with PCR. It is noteworthy that some H. pylori positive samples were also positive for Caga.Keywords
1. Introduction
Helicobacter pylori are spiral-shaped, microaerophilic, Gram-negative bacteria, and the main cause of gastroduodenal diseases (1). The prevalence of H. pylori infection in some developing countries is more than 80% and below 20% in some developed countries (2). Man is a major reservoir of the bacteria and colonization remains for a lifetime unless treated. The mode of transmission of H. pylori is from person to person with 2 route of transmission being proposed: fecal-oral, oral-oral, and/or stomach-oral routes. Studies suggest that the infection transmits through saliva and dental plaque, and normal and diarrheal stools (3). However, several studies show that low standard of living and crowded households are the major risk factors for contracting H. pylori infection (4, 5). Researches suggest environmental reservoirs such as food, water and domestic animal for H. pylori. To support this hypothesis, numerous studies are carried out to report the presence of H. pylori in such environments (6-10).
The presence of Helicobacter spp. is investigated in natural environmental waters, including ground water, fresh water streams, and off-shore marine waters using molecular technique, but it is rarely isolated from these kinds of samples (11-16). Since H. pylori have the propensity for exhibiting 2 forms: spiral and coccoid. The spiral-shaped form is metabolically active while the coccoid shape is formed under stressful conditions (17, 18). Some authors consider the coccoid form as dormant and viable (17, 19-21). Morphological change from spiral to coccoid makes the organism non-culturable (22). The coccoid form can change into the spiral form under certain conditions and this supposition increases the risk of H. pylori transmission through water. Studies show that the coccoid form of H. pylori, with a smaller amount of DNA and mRNA relative to the spiral form, has oxidative metabolism and respiration (18, 23). Therefore, water can play a major role in H. pylori transmission and the bacteria can survive as long as 20 days in distilled water, while their pathogenic ability remains (24).
The survival of H. pylori in water may also depend on specific factors such as the presence of free living amoeba and zooplanktons (25). Some studies show that H. pylori can be incorporated into the biofilm, which is an important factor for successful survival in the aquatic environment (26, 27). For successful long-term colonization in the human stomach, H. pylori harbors a set of bacterial virulence determinants; Cytotoxin-associated antigen (cagA) is 1 of widely disseminated genes with 90% prevalence among Asian population and is believed to increase the risk of gastric cancer (28). Therefore, the presence of this gene in water could be a risk factor for cancer development.
In the current study, the following hypothesis was devised based on the author’s previous studies: the presence of H. pylori was detected using PCR method targeting ureC gene. Therefore, the current study was used to determine if the analytical method commonly used for other bacteria can be used to evaluate and estimate the presence of H. pylori in water. Also, in the current study, cagA and 16s rRNA genes were targeted by PCR, and ureC for the Loop-mediated isothermal amplification (LAMP) method. For this purpose, a highly specific PCR method was developed and then the obtained results were compared with those of LAMP to evaluate the sensitivity and specificity of the LAMP method to detect H. pylori DNA in water or human samples; where a very high rate of H. pylori infection is recently shown (29, 30). In addition, samples were collected and analyzed with rigorous controls for false positive or negative results.
2. Methods
2.1. Sample Collection and Preparation
As mentioned in the authors’ previous study, samples were collected from urban tap and well water sources in Kermanshah from June to November 2012 (12). Kermanshah is a mountainous city with mild climate, and is the capital city of the province located in the Western part of Iran (31). The drinking water supplies Kermanshah come from 133 wells and Gavshan dam, which is connected to 21 reservoir tanks and distributed around the city (32).
2.2. DNA Preparation
The drinking water samples were obtained from taps of all water supplies. For each sample, 1 liter of water was obtained and filtered through 0.45 μm nitrocellulose filters (12). Finally, the DNA of the filtered bacteria was extracted using QIAAmp Mini DNA kit (Qiagen, Germany), according to the manufacturer’s instructions. In the authors’ previous study, the presence of H. pylori was analyzed using only PCR. Here, to evaluate the presence of 16s rRNA, cagA, and ureC genes, PCR was amplified using specific primers; LAMP targeted species specific ureC gene on the DNAs extracted from the samples.
2.3. Primer Design
The primers used in PCR in the current study were retrieved from previously published articles. Oligonucleotide primers targeting ureC (GenBank accession no. AE000511) for LAMP were designed using LAMP primer design software (http://primerexplorer.jp/e/index.html). The 2 outer primers were designated as forward outer primer (UreCF3) and backward outer primer (UreCB3). The inner primers were designated as Forward Inner Primer (UreC FIP) and Backward Inner Primer (UreC BIP). The loop primers were designated as forward and backward Loop Primers, (UreC LF) and (UreC LB), respectively. Complete sets of primers and their sequences are shown in Table 1.
Primers Used in the Current Study
| Primer | Sequence (5’ – 3’) | Position | Product Size | Reference |
|---|---|---|---|---|
| HP-UreCBIP | CTCGCCTCCAAAATTGGCTTGCGATTGGGGATAAGTTTG | The current study | ||
| HP-UreCFIP | GCATATCATTTTTAGCGATTACGCTCACTAACGCGCTCACTTG | |||
| HP-UreCB3 | TCCCAAGATTTGGAATTGAAG | |||
| HP-UreCF3 | GCTTACCTGCTTGCTTTC | |||
| HP-UreCLB | TCAATTGCATGCATTCGCTCA | |||
| HP-UreCLF | CAGGCGATGGTTTGGTGTG | |||
| ureC | ureCF: CAT CGC CAT CAA AAG CAA AG | 605 - 625 | 214 | (12) |
| ureCR: AGT TTA AGG ATC GTG TTA G | 798 - 819 | |||
| 16s rRNA | Rg; GCTATG ACG GGT ATC C | 276 - 291 | 400 | (33) |
| Fg: GAT TTTACC CCT ACA CCA | 681 - 698 | |||
| cagA | F1:GATAACAGGCAAGCTTTTGAGG | 349 | (34) | |
| B1:CTGCAAAAGATTGTTTGGCAGA |
2.4. PCR Assay
Optimal PCR reaction was carried out according to the procedure laid down in a previous article of the authors. In summary, the process was carried out using a final volume of 15 μL containing 20 mM Tris-HCl (Cinacolon, Iran) , 50 mM KCl, 200 μM dNTP mix (Cinacolon, Iran), 1.5 mM MgCl2, 0.5 μM of each forward and reverse primers (Takapouzist, Iran), 1 unit Taq DNA polymerase (Cinacolon, Iran), and 5 μL of template DNA. Initial denaturation of the target DNA was at 95°C for 5 minutes as well as 214, 422, and 349 bp target sequences were amplified in the reaction mix through 35 cycles as follows: 92°C for 30 seconds, 55°C for 30 seconds (ureC), 58°C for 30 seconds (cagA), 45°C for 30 seconds (16s rRNA gene) and 72°C for 30 seconds, followed by 72°C for 5 minutes. Electrophoresis through 1% agarose gel and staining with ethidium bromide were done on PCR product.
2.5. Loop-Mediated Isothermal Amplification Assay
Loop-mediated isothermal amplification was carried out in a total volume of 30 μL reaction volume. The LAMP system contained 1.6 μM of each FIP and BIP, 0.2 μM of each F3 and B3, 0.8 μM of each LF and BF, 8 U Bst polymerase, 3 μL of genomic DNA, 2 mM each dNTP (TransGen Biotech), 0.8 M betaine (Sigma, St. Louis, MO, USA), 20 mM Tris-HCl, 10 mM KCL, 10 mM (NH4)2SO4, 12 mM MgSO4 that was mixed and mixture transferred to microtubes. The reaction mixture was heated at 95°C for 3 minutes in thermocycler and then chilled on ice, 8 U Bst polymerase (New England Biolabs, USA) were added followed by incubation at 65°C for 60 minutes, and was heated at 96°C for 2 minutes and cooled at 4°C for 5 minutes to terminate the reaction (35).
2.6. Analysis of LAMP Products
The LAMP products were centrifuged for 3 minutes at 3000 rpm, positive samples can be detected through observable magnesium sulfide (MgSO4) with sediment at the bottom of microtubes. Negative LAMP products with no observable sediment were subjected to electrophoresis through 1.5% agarose gels then visualized under UV light after staining with ethidium bromide (Cinacolon, Iran) to prove negativity.
2.7. Determination of Primers Specificity in LAMP Reaction
To determine the specificity of LAMP primers, the primers were set up in a LAMP reaction with the DNA of other bacterial species such as Citrobacter, Campylobacter, Klebsiella, Yersinia, Shigella, and Pseudomonas, in addition to Enterococcus faecalis and Salmonella typhi.
2.8. Determination of Analytical Sensitivity
Furthermore, analytical sensitivity was carried out to detect the sensitivity of LAMP. Here, a suspension of 3 day old culture of H. pylori was prepared in a PBS buffer and the number of H. pylori was counted using Petroff-Hasser counting chamber. The average number of counted bacteria was used as the basis for multiple suspensions, which was approximately 1 to 100,000 bacteria per liter of water. The sensitivity of LAMP was evaluated with a detection limit and after DNA extraction of H. pylori, the concentration of DNA was measured 3 times using the Nonodrop spectrophotometer device and the average result was considered as a base to prepare serial dilution with concentrations of 10 ng/reaction to 0.01 fg/reaction.
2.9. Detection Limit
The sensitivity, specificity, and negative and positive predicting values of LAMP method, in line with PCR, were measured.
2.10. Statistical Analysis
Data were analyzed by SPSS software version 16. To calculate the degree of agreement between the 2 methods, Kappa coefficient was used. The gold standard for H. pylori detection was PCR method. Loop-mediated isothermal amplification sensitivity and specificity, negative and positive predicting values and its agreement with PCR was calculated using Table 2.
Specificity and Sensitivity Calculation
| Gold Standard (PCR) | |||
|---|---|---|---|
| Positive | Negative | ||
| LAMP | Positive | a (True positives) | b (False - positives) |
| Negative | c (False - negatives) | d (True negatives) | |
3. Results
3.1. PCR and LAMP Results
In a total of 60 drinking water samples, 16 (26.67%) were positive for ureC by PCR, 20 (33.33%) were positive for ureC by LAMP, and 37 (61.67%) were positive for 16s rRNA by PCR. Overall, detection rate based on ureC gene by PCR and LAMP was 26%.
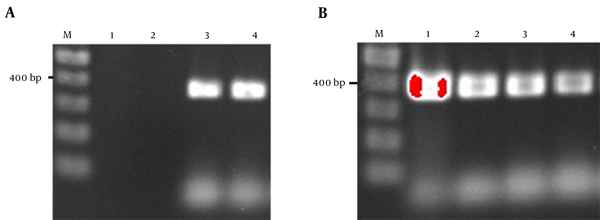
The prevalence of cagA gene among ureC positive samples was 3/16 (18.75%). But among 37 samples positive for 16s rRNA, 5 samples (13.51%) were also positive for cagA gene. The overall detection rate of cagA was 10% (6/60). Figure 1A and 1B show the PCR electrophoresis products of 16s rRNA and cagA genes.
Targeted 16s rRNA
3.2. Loop-Mediated Isothermal Amplification Specificity
The evaluation of primers specificity in LAMP reaction for ureC gene of H. pylori was done to decline any complementary relevancy between these primers and other bacterial genes in water. The results showed that the primers designed for ureC to be used in LAMP reaction were 100% specific for H. pylori.
3.3. Loop-Mediated Isothermal Amplification Sensitivity
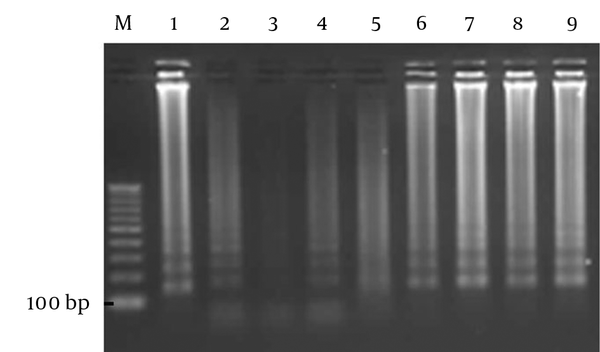
The analytical sensitivity of the LAMP method was 50 bacteria per liter with observable sediment, but after electrophoresis, the sensitivity improved to 1 bacterium per liter. The ability of the LAMP reaction to detect H. pylori, using pure DNA of the bacterium, was 10 fg/reaction (Figure 2). In the current study, the sensitivity, specificity, negative and positive predicting values of the LAMP to detect H. pylori in water samples were 100.00%, 90.91%, 100.00%, and 80.00%, respectively and the efficacy of the LAMP was 100% in comparison with that of PCR, which is the gold standard. Kappa coefficient and agreement between LAMP and PCR were 0.84 and 93%, respectively. The agreement between the 2 tests was excellent and can be used parallel to PCR.
The Electrophoresis of LAMP Product from Different Suspensions of H. pylori
4. Discussion
Adequate knowledge regarding the reservoirs and modes of transmitting H. pylori could help to explain the high prevalence of the bacteria. The incidence of H. pylori is high in the developing countries (90%), whereas in the industrialized countries, the figure is lower (50%) and tends to decrease. Childhood is the critical period for infection, and transmission usually occurs from person to person (2, 10, 36, 37). In an endemic area, a common source of infection is suspected (38).
Data of the present study showed that H. pylori can be detected in Kermanshah municipal tap water and the consumption of such water could be associated with gastric colonization of the organism. These findings were in line with those of the previous studies of the authors, but further investigation is required to determine whether the organism is viable or not (39). These findings also confirmed the previous observations in Isfahan, Iran (13), Colombia (40), Peru (41), Mexico (42), England (43), Sweden (38, 39, 41, 42, 44), Japan (45), and the United States (46). The high prevalence of H. pylori detected in drinking water samples strengthens the evidence of H. pylori transmission through drinking water. Considering that the cagA is associated with increased virulence and risk of peptic ulcer and cancer, the present study was the first to report on the presence of cagA in drinking water samples. A prevalence of 13.51% of this gene in drinking water is an alarming situation. In a similar study carried out in Pakistan, the prevalence of 16s rRNA and cagA were 40% and 0, respectively (47).
The result obtained for 16s rRNA was considerably high, 61.67% and 25% (15/60) of 16s rRNA positive samples were negative for the LAMP of ureC gene. This indicates the likely presence of other Helicobacter spp. other than H. pylori in the water samples. Poor sanitation of water and allowing domestic animals, which could be a carrier of non-pylori Helicobacter species to roam near water supplies, lead to water contamination (48-52). Another probability is the presence of H. pylori that has lost its pathogenicity genes (53).
The current study was also the first to report on the possible existence of Helicobacter ssp. in water samples. Non-pylori Helicobacter species are associated with some human diseases and could exacerbate some situations such as inflammatory bowel disease (IBD), and hepatocellular carcinoma (HCC) (54-56). Among the ureC positive samples, 18.75% were positive for the cagA gene and the overall detection of cagA gene was 13.51%. No study is carried out to detect cagA gene in water, but in a previous study on gastric biopsies, in terms of prevalence, cagA was 84.4% (30).
The cagA is a 40 kbp gene located in the cag pathogenicity island (PaI) of the H. pylori chromosome (57). It is shown that the presence of cagA gene is associated with peptic ulcer disease (58), atrophic gastritis (59), and gastric adenocarcinoma (60). The cagA positive strains are more virulent than other strains (61). The presence of cagA gene in water sample could be a potential risk for cancer development in Kermanshah, Iran. All the mentioned previous studies were based on the PCR of ureC gene or 16s rRNA gene, but in the current study 2 methods of detection were considered; PCR and LAMP. In addition, various genes were employed as a target of amplification, which caused an increase in detection accuracy.
It is noteworthy that it was the first study to report on the use of LAMP reaction to detect H. pylori in water samples.
Loop-mediated isothermal amplification is easy to perform if the appropriate primers are prepared, which requires only 6 pairs of primers, DNA polymerase, a bain-marie bath, and a thermocycler for reaction. Loop-mediated isothermal amplification is 10-100-fold more analytically sensitive than PCR. Compared to the other amplification methods, the DNA amplification reaction in LAMP method is carried out under isothermal condition and the efficiency of the amplification is higher and, the detection limit is lower (62, 63). The analytical specificity of the LAMP is attributed to 6 sets of primers that recognize 8 distinct regions on the target DNA. The amplified products can be also confirmed using sequencing or digestion with restriction enzyme (64-66). Since the LAMP method is much more analytically sensitive than PCR, therefore, more positive results are obtained in the reactions.
Considering the fact that cagA is associated with increased virulence, risk of peptic ulcer and cancer, the high prevalence of H. pylori and the presence of cagA gene in drinking water is fast becoming an alarming situation. In the current study, 25% of samples were positive for non-pylori Helicobacter species. However, non-pylori Helicobacter species are linked with chronic infection of the intestinal and hepatobiliary tract. They also disturb immune responses of the intestinal epithelial cells by modulating its inflammatory response, which increases the risk of bacterial infection in the intestine. The contamination of water by these bacteria could be a potential risk to develop some gastrointestinal diseases.
Acknowledgements
References
-
1.
Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;113(3):321-33. [PubMed ID: 14755326]. https://doi.org/10.1172/JCI20925.
-
2.
Bardhan PK. Epidemiological features of Helicobacter pylori infection in developing countries. Clin Infect Dis. 1997;25(5):973-8. [PubMed ID: 9402340].
-
3.
Akcan Y, Ersan S, Alper M, Bicik Z, Aytug N. The transmission of Helicobacter pylori via exposure to common sources outweighs the person-to-person contact among spouses in developing countries. Am J Gastroenterol. 2000;95(1):317-9. [PubMed ID: 10638617]. https://doi.org/10.1111/j.1572-0241.2000.01722.x.
-
4.
Brown LM, Thomas TL, Ma JL, Chang YS, You WC, Liu WD, et al. Helicobacter pylori infection in rural China: demographic, lifestyle and environmental factors. Int J Epidemiol. 2002;31(3):638-45. [PubMed ID: 12055167].
-
5.
Graham DY, Adam E, Reddy GT, Agarwal JP, Agarwal R, Evans DJ, et al. Seroepidemiology of Helicobacter pylori infection in India. Comparison of developing and developed countries. Dig Dis Sci. 1991;36(8):1084-8. [PubMed ID: 1864201].
-
6.
Karita M, Teramukai S, Matsumoto S. Risk of Helicobacter pylori transmission from drinking well water is higher than that from infected intrafamilial members in Japan. Dig Dis Sci. 2003;48(6):1062-7. [PubMed ID: 12822863].
-
7.
Klein PD, Graham DY, Gaillour A, Opekun AR, Smith EO. Water source as risk factor for Helicobacter pylori infection in Peruvian children. Gastrointestinal Physiology Working Group. Lancet. 1991;337(8756):1503-6. [PubMed ID: 1675369].
-
8.
Perez-Perez GI, Rothenbacher D, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9 Suppl 1:1-6. [PubMed ID: 15347299]. https://doi.org/10.1111/j.1083-4389.2004.00248.x.
-
9.
Vale FF, Vitor JM. Transmission pathway of Helicobacter pylori: does food play a role in rural and urban areas? Int J Food Microbiol. 2010;138(1-2):1-12. [PubMed ID: 20122750]. https://doi.org/10.1016/j.ijfoodmicro.2010.01.016.
-
10.
van Duynhoven YT, de Jonge R. Transmission of Helicobacter pylori: a role for food? Bull World Health Organ. 2001;79(5):455-60. [PubMed ID: 11417041].
-
11.
Adams BL, Bates TC, Oliver JD. Survival of Helicobacter pylori in a natural freshwater environment. Appl Environ Microbiol. 2003;69(12):7462-6. [PubMed ID: 14660399].
-
12.
Amirhooshang A, Ramin A, Ehsan A, Mansour R, Shahram B. High frequency of Helicobacter pylori DNA in drinking water in Kermanshah, Iran, during June-November 2012. J Water Health. 2014;12(3):504-12. [PubMed ID: 25252354]. https://doi.org/10.2166/wh.2013.150.
-
13.
Bahrami AR, Rahimi E, Ghasemian Safaei H. Detection of Helicobacter pylori in city water, dental units' water, and bottled mineral water in Isfahan, Iran. ScientificWorldJournal. 2013;2013:280510. [PubMed ID: 23606812]. https://doi.org/10.1155/2013/280510.
-
14.
Baker KH, Hegarty JP. Presence of Helicobacter pylori in drinking water is associated with clinical infection. Scand J Infect Dis. 2001;33(10):744-6. [PubMed ID: 11728039].
-
15.
Benson JA, Fode-Vaughan KA, Collins ML. Detection of Helicobacter pylori in water by direct PCR. Lett Appl Microbiol. 2004;39(3):221-5. [PubMed ID: 15287865]. https://doi.org/10.1111/j.1472-765X.2004.01555.x.
-
16.
Luzza F, Imeneo M, Maletta M, Paluccio G, Giancotti A, Perticone F, et al. Seroepidemiology of Helicobacter pylori infection and hepatitis A in a rural area: evidence against a common mode of transmission. Gut. 1997;41(2):164-8. [PubMed ID: 9301493].
-
17.
Nilius M, Strohle A, Bode G, Malfertheiner P. Coccoid like forms (CLF) of Helicobacter pylori. Enzyme activity and antigenicity. Zentralbl Bakteriol. 1993;280(1-2):259-72. [PubMed ID: 8280950].
-
18.
She FF, Lin JY, Liu JY, Huang C, Su DH. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J Gastroenterol. 2003;9(3):516-20. [PubMed ID: 12632509].
-
19.
Kusters JG, Gerrits MM, Van Strijp JA, Vandenbroucke-Grauls CM. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect Immun. 1997;65(9):3672-9. [PubMed ID: 9284136].
-
20.
Mizoguchi H, Fujioka T, Nasu M. Evidence for viability of coccoid forms of Helicobacter pylori. J Gastroenterol. 1999;34 Suppl 11:32-6. [PubMed ID: 10616763].
-
21.
Ren Z, Pang G, Musicka M, Dunkley M, Batey R, Beagley K, et al. Coccoid forms of Helicobacter pylori can be viable. Microbios. 1999;97(388):153-63. [PubMed ID: 10413871].
-
22.
Cellini L, Allocati N, Angelucci D, Iezzi T, Di Campli E, Marzio L, et al. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol. 1994;38(11):843-50. [PubMed ID: 7898382].
-
23.
Saito N, Konishi K, Sato F, Kato M, Takeda H, Sugiyama T, et al. Plural transformation-processes from spiral to coccoid Helicobacter pylori and its viability. J Infect. 2003;46(1):49-55. [PubMed ID: 12504609].
-
24.
Shahamat M, Mai U, Paszko-Kolva C, Kessel M, Colwell RR. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl Environ Microbiol. 1993;59(4):1231-5. [PubMed ID: 8489232].
-
25.
Cellini L, Del Vecchio A, Di Candia M, Di Campli E, Favaro M, Donelli G. Detection of free and plankton-associated Helicobacter pylori in seawater. J Appl Microbiol. 2004;97(2):285-92. [PubMed ID: 15239694]. https://doi.org/10.1111/j.1365-2672.2004.02307.x.
-
26.
Costerton JW, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176(8):2137-42. [PubMed ID: 8157581].
-
27.
Linke S, Lenz J, Gemein S, Exner M, Gebel J. Detection of Helicobacter pylori in biofilms by real-time PCR. Int J Hyg Environ Health. 2010;213(3):176-82. [PubMed ID: 20427237]. https://doi.org/10.1016/j.ijheh.2010.03.006.
-
28.
Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15(3):306-16. [PubMed ID: 24629337]. https://doi.org/10.1016/j.chom.2014.02.008.
-
29.
Bakhtiari S, Alvandi A, Pajavand H, Navabi J, Najafi F, Abiri R. Development and Diagnostic Evaluation of Loop-Mediated Isothermal Amplification Using a New Gene Target for Rapid Detection of Helicobacter pylori. Jundishapur J Microbiol. 2016;9(5). ee28831. [PubMed ID: 27540449]. https://doi.org/10.5812/jjm.28831.
-
30.
Pajavand H, Alvandi A, Mohajeri P, Bakhtyari S, Bashiri H, Kalali B, et al. High Frequency of vacA s1m2 Genotypes Among Helicobacter pylori Isolates From Patients With Gastroduodenal Disorders in Kermanshah, Iran. Jundishapur J Microbiol. 2015;8(11). ee25425. [PubMed ID: 26862378]. https://doi.org/10.5812/jjm.25425.
-
31.
Climatology and geography of Kermanshah province. 2012. Available from: www.kermanshahmet.ir.
-
32.
Kermanshah water and sewage company Kermanshah. 2012. Available from: http://www.abfaksh.ir/ab/.
-
33.
Stolte M, Wellens E, Bethke B, Ritter M, Eidt H. Helicobacter heilmannii (formerly Gastrospirillum hominis) gastritis: an infection transmitted by animals? Scand J Gastroenterol. 1994;29(12):1061-4. [PubMed ID: 7886392].
-
34.
Chen XJ, Yan J, Shen YF. Dominant cagA/vacA genotypes and coinfection frequency of H. pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases. Chin Med J (Engl). 2005;118(6):460-7. [PubMed ID: 15788126].
-
35.
Amiri N, Abiri R, Eyvazi M, Zolfaghari MR, Alvandi A. The frequency of Helicobacter pylori in dental plaque is possibly underestimated. Arch Oral Biol. 2015;60(5):782-8. [PubMed ID: 25766471]. https://doi.org/10.1016/j.archoralbio.2015.02.006.
-
36.
Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10(4):720-41. [PubMed ID: 9336670].
-
37.
Farthing MJ. Helicobacter pylori infection: an overview. Br Med Bull. 1998;54(1):1-6. [PubMed ID: 9604425].
-
38.
Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22(2):283-97. [PubMed ID: 11218379].
-
39.
Piqueres P, Moreno Y, Alonso JL, Ferrus MA. A combination of direct viable count and fluorescent in situ hybridization for estimating Helicobacter pylori cell viability. Res Microbiol. 2006;157(4):345-9. [PubMed ID: 16380234]. https://doi.org/10.1016/j.resmic.2005.09.003.
-
40.
Goodman KJ, Correa P, Tengana Aux HJ, Ramirez H, DeLany JP, Guerrero Pepinosa O, et al. Helicobacter pylori infection in the Colombian Andes: a population-based study of transmission pathways. Am J Epidemiol. 1996;144(3):290-9. [PubMed ID: 8686698].
-
41.
Hulten K, Han SW, Enroth H, Klein PD, Opekun AR, Gilman RH, et al. Helicobacter pylori in the drinking water in Peru. Gastroenterology. 1996;110(4):1031-5. [PubMed ID: 8612990].
-
42.
Mazari-Hiriart M, Lopez-Vidal Y, Calva JJ. Helicobacter pylori in water systems for human use in Mexico City. Water Sci Technol. 2001;43(12):93-8. [PubMed ID: 11464777].
-
43.
Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, et al. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol. 2004;97(4):690-8. [PubMed ID: 15357718]. https://doi.org/10.1111/j.1365-2672.2004.02360.x.
-
44.
Hulten K, Enroth H, Nystrom T, Engstrand L. Presence of Helicobacter species DNA in Swedish water. J Appl Microbiol. 1998;85(2):282-6. [PubMed ID: 9750301].
-
45.
Horiuchi T, Ohkusa T, Watanabe M, Kobayashi D, Miwa H, Eishi Y. Helicobacter pylori DNA in drinking water in Japan. Microbiol Immunol. 2001;45(7):515-9. [PubMed ID: 11529557].
-
46.
Hegarty JP, Dowd MT, Baker KH. Occurrence of Helicobacter pylori in surface water in the United States. J Appl Microbiol. 1999;87(5):697-701. [PubMed ID: 10594710].
-
47.
Khan A, Farooqui A, Kazmi SU. Presence of Helicobacter pylori in drinking water of Karachi, Pakistan. J Infect Dev Ctries. 2012;6(3):251-5. [PubMed ID: 22421606].
-
48.
Boomkens SY, Kusters JG, Hoffmann G, Pot RG, Spee B, Penning LC, et al. Detection of Helicobacter pylori in bile of cats. FEMS Immunol Med Microbiol. 2004;42(3):307-11. [PubMed ID: 15477044]. https://doi.org/10.1016/j.femsim.2004.06.002.
-
49.
Engstrand L. Helicobacter in water and waterborne routes of transmission. Symp Ser Soc Appl Microbiol. 2001;(30):80S-4S. [PubMed ID: 11422563].
-
50.
Fox JG, Batchelder M, Marini R, Yan L, Handt L, Li X, et al. Helicobacter pylori-induced gastritis in the domestic cat. Infect Immun. 1995;63(7):2674-81. [PubMed ID: 7790084].
-
51.
Neiger R, Simpson KW. Helicobacter infection in dogs and cats: facts and fiction. J Vet Intern Med. 2000;14(2):125-33. [PubMed ID: 10772482].
-
52.
Van den Bulck K, Decostere A, Baele M, Driessen A, Debongnie JC, Burette A, et al. Identification of non-Helicobacter pylori spiral organisms in gastric samples from humans, dogs, and cats. J Clin Microbiol. 2005;43(5):2256-60. [PubMed ID: 15872252]. https://doi.org/10.1128/JCM.43.5.2256-2260.2005.
-
53.
Covacci A, Falkow S, Berg DE, Rappuoli R. Did the inheritance of a pathogenicity island modify the virulence of Helicobacter pylori? Trends Microbiol. 1997;5(5):205-8. [PubMed ID: 9160510]. https://doi.org/10.1016/S0966-842X(97)01035-4.
-
54.
Hansen R, Berry SH, Mukhopadhya I, Thomson JM, Saunders KA, Nicholl CE, et al. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One. 2013;8(3). ee58825. [PubMed ID: 23554935]. https://doi.org/10.1371/journal.pone.0058825.
-
55.
Powers-Fletcher MV, Couturier MR. Non-Helicobacter pylori Helicobacter Species Associated with Human Disease: a Primer for the Clinical Microbiology Laboratory. Clin Microbiol Newsletter. 2015;37(12):93-101. https://doi.org/10.1016/j.clinmicnews.2015.05.004.
-
56.
Yang J, Ji S, Zhang Y, Wang J. Helicobacter hepaticus infection in primary hepatocellular carcinoma tissue. Singapore Med J. 2013;54(8):451-7. [PubMed ID: 24005452].
-
57.
Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43(8):3906-16. [PubMed ID: 16081930]. https://doi.org/10.1128/JCM.43.8.3906-3916.2005.
-
58.
Peek RM Jr, Miller GG, Tham KT, Perez-Perez GI, Zhao X, Atherton JC, et al. Heightened inflammatory response and cytokine expression in vivo to cagA+ Helicobacter pylori strains. Lab Invest. 1995;73(6):760-70. [PubMed ID: 8558837].
-
59.
Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40(3):297-301. [PubMed ID: 9135515].
-
60.
Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111-5. [PubMed ID: 7743510].
-
61.
Crabtree JE, Taylor JD, Wyatt JI, Heatley RV, Shallcross TM, Tompkins DS, et al. Mucosal IgA recognition of Helicobacter pylori 120 kDa protein, peptic ulceration, and gastric pathology. Lancet. 1991;338(8763):332-5. [PubMed ID: 1677696].
-
62.
Hong TC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(5):1956-61. [PubMed ID: 15131154].
-
63.
Parida M, Horioke K, Ishida H, Dash PK, Saxena P, Jana AM, et al. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J Clin Microbiol. 2005;43(6):2895-903. [PubMed ID: 15956414]. https://doi.org/10.1128/JCM.43.6.2895-2903.2005.
-
64.
Aryan E, Makvandi M, Farajzadeh A, Huygen K, Alvandi AH, Gouya MM, et al. Clinical value of IS6110-based loop-mediated isothermal amplification for detection of Mycobacterium tuberculosis complex in respiratory specimens. J Infect. 2013;66(6):487-93. [PubMed ID: 23466595]. https://doi.org/10.1016/j.jinf.2013.02.005.
-
65.
Aryan E, Makvandi M, Farajzadeh A, Huygen K, Bifani P, Mousavi SL, et al. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiol Res. 2010;165(3):211-20. [PubMed ID: 19515543]. https://doi.org/10.1016/j.micres.2009.05.001.
-
66.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. [PubMed ID: 10871386].