Abstract
Background:
Lactobacillus plantarum is a widespread probiotic bacterium. Many plasmids from L. plantarum have been identified to encode some important phenotypic traits including carbohydrate metabolism, bacteriocin synthesis, and exopolysaccharide production.Objectives:
The aim of this study was to identify and characterize the native plasmids from L. plantarum BM4.Methods:
Lactobacillus plantarum BM4 was isolated from fermented meat in Guangxi Province, China, and characterized by 16S rRNA sequence. Four plasmids were isolated from L. plantarum BM4, sequenced, and characterized by the bioinformatics method. Moreover, the relative copy numbers of these plasmids were estimated using the droplet digital PCR method.Results:
Four plasmids, designated as pBM1, pBM2, pBM3, and pBM4, were isolated from L. plantarum BM4. By nucleotide sequencing, pBM1, pBM2, pBM3, and pBM4 were characterized as having sizes of 6069 bp, 7042 bp, 8131 bp, and 8892 bp, and G+C contents of 37.5%, 36.7%, 36.4%, and 34.5%, respectively. Nucleotide sequence analysis revealed 8, 10, 10, and 10 putative open reading frames (ORFs) for pBM1, pBM2, pBM3, and pBM4 plasmids, respectively. Based on sequence alignment, only pBM2 contained replication protein RepB and rep3, which contained a putative repeat origin of replication segment, indicating that the pBM2 belongs to the pUCL287 subfamily of theta-type replicons. Finally, the relative copy numbers of pBM1-4 were estimated to be 82, 24, 34, and 16, copies, respectively.Conclusions:
Four novel plasmids were isolated from L. plantarum BM4 and characterized. These backbones can potentially be developed for use as a cloning or expressing vectors in biotechnology applications.Keywords
1. Background
Lactobacillus plantarum is widely used in starter cultures in industrial food fermentation and it plays a beneficial role in health as a probiotic bacterium (1, 2). Lactobacillus plantarum species often harbor one or more natural plasmids (3). Many important properties of L. plantarum, such as resistance to phages, lactose catabolism, bacteriocins, and exopolysaccharides, are encoded by their plasmids (4). With the development of biology and genetics, characterization of the potential valuable tools of L. plantarum plasmids has become a hot area of research in recent years.
To date, many plasmids of L. plantarum have been sequenced, such as pLP9000 (9.3 kb) (5), pPB1 (2.9 kb) (6), and pLKS (2.0 kb) (7), and the number of sequenced plasmids have been steadily increasing. At least three different mechanisms of plasmid replication have been recognized, namely theta type, strand displacement, and rolling circle mode of replication (RCR), among which RCR is widespread in bacterial plasmids (8). Bacterial plasmids usually initiate replication by a plasmid-encoded protein, generically termed Rep (9).
In general, initiation of RCR needs recognition of the plasmid dso by the cognate Rep protein, involving the synthesis of single-strand DNA (ssDNA) intermediates (10). Based on sequence similarity in the Rep protein and double-stranded origin (dso), RCR plasmids can be grouped into several families, e.g., pT181, pE194/pLS1, pC194/pUB110, pSN2, and pMV158 (11). The theta type plasmids contain the rep gene and a replicon that consists of an origin of replication (ori) and an AT-rich region that primarily involves repeats as well as a binding site (12). Theta replicons comprise three key components, which are plasmid-encoded initiator Rep proteins, origins of replication, and host-encoded DNA polymerase I for nascent strand DNA synthesis (13). The ori of these lactococcal replicons consists of a set of short repeats (10 or 11 bp) and it is finished by large repeats (20 or 22 bp) adjacent to the promoter of the initiator gene (14). In addition, at least five classes of theta replicons have been identified according to the mechanism of initiation of DNA replication (15, 16).
2. Objectives
In this paper, four novel plasmids named pBM1, pBM1, pBM1, and pBM4 were isolated from L. plantarum BM4. The nucleotides of the four plasmids were sequenced and characterized, and the copy numbers of the four plasmids in the cell were determined by Droplet Digital PCR.
3. Methods
3.1. Bacterial Strains, Plasmids and Growth Conditions
The L. plantarum BM4 used in this study was isolated from fermented meat in Guangxi Province, China. Lactobacillus plantarum BM4 was cultured on Man-Rogosa-Sharpe (MRS) medium (LuQiao, China) at 37°C under anaerobic conditions. Escherichia coli DH5a was used as a cloning host and cultured in Luria-Bertani (LB) medium (AoBoXing, China) with vigorous shaking at 37°C. Plasmid pBluescript II SK (+) (Stratagene, USA) was used as a subcloning vector for sequencing. When needed, ampicillin (at a final concentration of 100 μg/mL) was added to the medium.
3.2. 16S rRNA analysis of L. plantarum BM4
The genome DNA was isolated from L. plantarum BM4 using the TIANamp Bacteria DNA Kit (Tiangen, China) according to the manufacturer’s instruction. The 16S rRNA gene sequences were amplified from L. plantarum BM4 genomic DNA using PCR technique (Bio-Rad, USA) with universal primers 27F and 1492R (Table 1). The amplified fragment was subjected to agarose gel electrophoresis (1% w/v), and sequenced (Sangon Biotech, China). The 16S rDNA gene sequences of Lactobacillus were collected from the Genbank database. MEGA 4.0 was used to perform multiple sequence alignments using the ClustalW method (17).
3.3. Plasmid Sequencing and Analysis
The plasmids were isolated from L. plantarum BM4 by the alkaline lysis method with a slight modification (18). Briefly, cells were harvested after overnight culture by centrifugation at 10,000 g for 5 minutes, and then washed in TES buffer (50 mM Tris-Cl, 30 mM EDTA, 25% Sucrose, pH 8.0). Lysozyme was subsequently added at the final concentration of 50 mg.mL-1, and the suspension was incubated at 37°C for 1 hour. Plasmid DNA was isolated using a TIANprep Mini Plasmid Kit (Tiangen, China). The extracted total plasmid DNA was subjected to the 1% (w/v) agarose electrophoresis analysis.
Plasmid DNA isolated from L. plantarum BM4 and pBluescript II SK (+) was digested by KpnI Restriction endonuclease (RE) according to the supplier’s instructions (Takara, China). The RE-digested plasmids were subsequently cloned into the pBluescript II SK (+) vector and transformed into E. coli DH5a. Positive clone was selected by Ampicillin agar (100 μg/mL). Plasmid DNA from positive E. coli was isolated following the TIANprep Mini Plasmid Kit according to the manufacturer’s instruction (Tiangen, China). The inserted fragment was sequenced with T7 and T3 promoter primers using Applied Biosystems Automated Sequencer (Sangon Biotech, China), and the complete nucleotide sequence was obtained through primer walking method.
Open reading frames (ORF) were predicted by the ORF finder program at the national center for biotechnology information (NCBI) site and FGENESB program at the Softberry site (http://linux1.softberry.com/berry.phtml). Alignments of conserved protein domains were retrieved from the conserved domain database (CDD) at the NCBI (19). The DNASTAR software package was employed to detect direct and inverted repeats. The promoter was predicted at the Softberry site (http://linux1.softberry.com/berry.phtml).
3.4. Copy Number Determination by Digital PCR
The L-lactate dehydrogenase gene (ldhL1) (Genbank: NC_004567.2) in L. plantarum WCFS1, which was identified as a chromosomally encoded single-copy gene, was used as the reference gene. PCR primers used in digital PCR were shown in Table 1. A single ddPCR reaction volume of 20 µL contained 10 µL 2x ddPCR supermix (Bio-Rad, USA), 2 µL primers (final concentration of 10 µM), 2 µL DNA template, and 6 µL ddH2O. Reaction mixtures were briefly mixed by vortexing while avoiding the formation of bubbles, micro-centrifuged for 20 sec, and then kept on ice until droplet generation. Samples were converted into droplets with the QX100 Droplet Digital PCR (ddPCR) system (Bio-Rad, USA) according to the manufacturer’s instruction. Next, the droplets were transferred from the droplet wells in the DG8 cartridges (Bio-Rad, USA) to a 96-well PCR plate and sealed for 5 seconds with a heat sealer. After that, amplification was performed in the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) under the following cycling conditions: 95ºC for 6 minutes followed by 40 cycles of 95°C for 15 seconds and 52°C for 45 seconds. After PCR, the plate was loaded onto the QX100 Droplet Digital reader (Bio-Rad, USA), which automatically reads the droplets from each well of the plate.
Primers Used in This Study
| Target | Sequence (5'-3') | Amplification Size, bp |
|---|---|---|
| 16S rRNA | 27F: AGAGTTTGATCCTGGCTCAG | 1465 |
| 1492R: TACGGCTACCTTGTTACGACTT | ||
| ldhL1 | ldh-F: CACCGTCTTCTAACTTGGCT | 152 |
| ldh-R: TCCTCGTTCCGTTGATGC | ||
| pMB1 | pBM1-F: TAGCACGATTTTGACCAG | 116 |
| pBM1-R: CACCAAGCGAAACTAACG | ||
| pMB2 | pBM2-F: TCTTATTAGATGGGCTATTTG | 190 |
| pBM2-R: GGATTATCAGAGGCAAGGT | ||
| pMB3 | pBM3-F: CAGCCGTTGACCTATTGC | 130 |
| pBM3-R: TTCGCTTGGTGTTTTGTTT | ||
| pMB4 | pBM4-F: TGCCAACGAGGAAAATCA | 111 |
| pBM4-R: AATCAACCAGACCACGGA |
3.5. Nucleotide Sequence Accession Number
Lactobacillus plantarum BM4 16S ribosomal DNA gene, partial sequence has been deposited in GenBank under Accession No. KP976095.1. The complete nucleotide sequences of pBM1-4 have been deposited in GenBank (Accession No: KT149387, KT149388, KT149389, KT149390).
3.6. Statistical analysis
Date analysis was performed using Quantasoft software (Bio-Rad, USA). The plasmid relative copy number to the chromosome was calculated based on the equation: PCN (plasmid copy number) = Np/Nc, where Np is the copy number per µL of the plasmid target gene, and NC is the copy number per µL of the control (chromosomal target gene) with the same template. The experiments were performed in triplicate, and the average values were recorded.
4. Results
4.1. Isolation and Identification of L. plantarum BM4
Lactobacillus strain BM4 was originally isolated from fermented meat. The 16S rDNA gene sequence analysis indicated that strain BM4 shows 99% homology with L. plantarum strains (Figure 1). Consequently, the Lactobacillus strain BM4 belonged to the L. plantarum species and therefore, it was designated as L. plantarum BM4.
A Phylogenetic Tree was Constructed with the MEGA Version 4 Program Using 16S rDNA Gene Sequences

4.2. Sequence Analysis of Plasmid pBM1-4
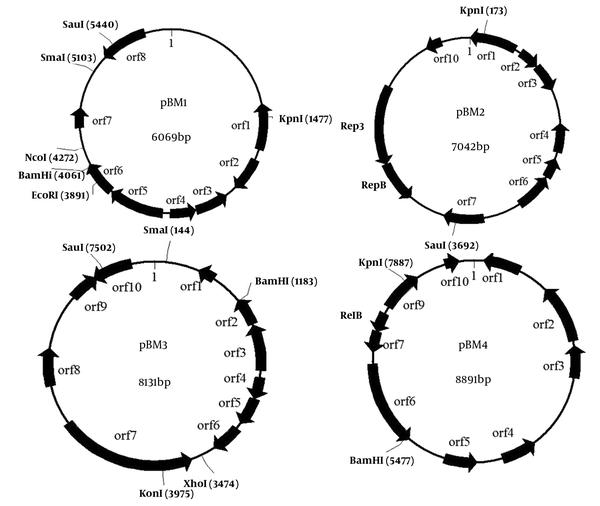
Lactobacillus plantarumBM4 harbored four plasmids, designated as pBM1, pBM2, pBM3, and pBM4, respectively. To sequence the plasmids, the four plasmids were digested with KpnI and then cloned into the vector pBluescript II SK (+). The sequence analysis results revealed that pBM1, pBM2, pBM3, and pBM4 were 6069 bp, 7042 bp, 8131 bp, and 8891 bp in length and possessed average G + C contents of 37.5%, 36.7%, 36.4%, and 34.5%, respectively. All open reading frames larger than 40 amino acids and their potential functions have been predicted at NCBI in the GenBank database. Physical maps of plasmids pBM1-4 are shown in Figure 2. We predicted 8 ORFs on pBM1, and 10 ORFs on pBM2, pBM3, and pBM4.
Physical Maps of Plasmid pBM1-4
4.3. Rep Proteins in Plasmid pBM2
According to the sequence analysis, among the four plasmids isolated from L. plantarum BM4, only pBM2 encoded a plasmid probable replication protein (repB) and an initiator replication family protein (repA), and the two genes overlapped in pBM2. Homology analysis results are shown in Figure 3. The repA protein (GenBank: ALO75838.1) of pMB2 shares 99% identity with the rep protein (GenBank: BAN08206.1) of plasmid pKB290-8, 80% identity with the repA protein (GenBank:CAA53278.1) of plasmid pUCL287, 86% identity with the repA protein (GenBank: NP_862285.1) of plasmid pMD5057, 78% identity with the repA protein (GenBank: NP_862269.1) of plasmid pRV500, 81% identity with the repB protein (GenBank: NP_857600.1) of plasmid pSMB74, and 78% identity with the initiation protein repA (GenBank: YP_002567764.1) of plasmid pRS5, all of which belong to the theta-replicating group.
In addition, supposed origins of replication (termed ori) are predicted upstream of repA (sites: 5749-6000) according to the structure feature, which consists of three direct 11-bp repeats (three times) and four 22-bp iterons, tandemly repeated (Figure 4). Moreover, Orf8 of pBM2 encoded a putative repB protein (GenBank: ALO75837.1) of 171 amino acids and shared 84% identity with the repB protein (GenBank: BAN08207.1) of plasmid pKB290-8 and 65% identity with the replication protein (GenBank: NP_857601.1) of plasmid pSMB74.
Amino Acid Sequence Alignment of the orf9-Coding Protein (RepA) of pBM2 with related plasmids RepA of PMD5057 (GenBank: AAN40880.1), RepA of pRS5 (GenBank: YP_002567764.1), RepA of pRV500 (GenBank: AAN61991.1), RepA of PUCL287 (GenBank: CAA53278.1), RepB of pSMB74 (GenBank: AAP55632.1).

Structure of the Predicted Origin of Replication in Plasmid pBM2

4.4. Other ORFS in Plasmid
All four plasmids contain a similar INT_C_like_3 Integrase protein of 195aa (orf5 in pBM1, orf1 in pBM2, orf3 in pBM3, and orf9 in pBM4), share over 90% identity with integrase of plasmids from Lactobacillus pentosus IG1 and Lactobacillus pentosus MP-10, and a transposase protein of 167aa (orf8 in pBM1, orf7 in pBM2, orf10 in pBM3, and orf4 in pBM4), share over 98% identity with transposase of plasmids from L. plantarum subsp. plantarum P-8. The four integrase proteins of pBM1-4 belong to a superfamily of DNA breaking/re-joining enzymes and might play a role in plasmid multimer resolution. Orf3 and orf4 in pBM1, as well as orf4 and orf5 in pBM2, encode a toxin-antitoxin (TA) plasmid maintenance system. Orf3-encoding protein in pBM1 and orf5 encoding protein in pBM3 belong to plasmid stabilization system protein. Orf2-encoding protein in pBM2 belongs to antidote-toxin recognition MazE. MazE is the antidote to the toxin MazF of E. coli, which regulates the prokaryotic chromosomal addiction module. Orf3 in pBM2 encodes a PemK-like protein, which is an inhibitor for growing in E. coli known to bind to the promoter region of the Pem operon, auto-regulating synthesis. No obvious replication protein, double-strand origin, or single-strand origin was found in plasmids pBM1, pBM3, or pBM4. Therefore, our data do not support their replication mechanism. The detailed characteristics of pBM1-4 are shown in Table 2.
Putative Genes and Their Products, Deduced from the Plasmid Nucleotide Sequences
| ORF | Position in Nucleotide Sequence, bp | G + C | Protein | Closest Relative (length, Level of Amino Acid Identity, Micro-Organism) | Accession no. | ||
|---|---|---|---|---|---|---|---|
| 5’ | 3’ | % | Length, aa | Molecular mass, Da | |||
| pBM1 | |||||||
| orf1 | 1820 | 1332 | 37.01 | 162 | 18640.6 | Hypothetical protein (162aa, 97%, Lactobacillus plantarum) | WP_015063543.1 |
| orf2 | 1933 | 2295 | 36.09 | 120 | 13825.7 | Transcriptional regulator (112aa, 96%, Lactobacillus plantarum) | WP_011031954.1 |
| orf3 | 2772 | 2416 | 36.97 | 118 | 13847.5 | Addiction module toxin RelE (plasmid) (118aa, 99, %Lactobacillus plantarum HFC8) | ALG27536.1 |
| Addiction module toxin, RelE/StbE family (plasmid) (118aa, 98%, Lactobacillus buchneri NRRL B-30929) | AEB74676.1 | ||||||
| orf4 | 3050 | 2772 | 35.48 | 92 | 10708.7 | Prevent-host-death family protein (93aa, 100%, Lactobacillus brevis) | WP_042253922.1 |
| Antitoxin (plasmid) (92aa, 98%, Lactobacillus plantarum HFC8) | ALG27537.1 | ||||||
| orf5 | 3127 | 3714 | 40.65 | 195 | 22480.7 | Integrase (195aa, 99%, Lactobacilluspentosus IG1) | CCC15493.1 |
| Integrase (195aa, 100%, Lactobacillus plantarum HFC8) | ALG27531.1 | ||||||
| orf6 | 3727 | 4086 | 35.28 | 119 | 14235.3 | Hypothetical protein (119aa, 100%, Lactobacillus plantarum) | WP_016511831.1 |
| Hypothetical protein (119aa, 98%, Lactobacillus brevis) | WP_015474693.1 | ||||||
| orf7 | 4483 | 4695 | 31.46 | 70 | 8288.3 | Hypothetical protein (70aa, 99%, Lactobacillus versmoldensis) | WP_010625611.1 |
| Hypothetical protein (70aa, 97%, Lactobacillus plantarum) | WP_027821986.1 | ||||||
| orf8 | 5786 | 5283 | 46.63 | 167 | 18712.4 | Hypothetical protein (167aa, 99%, Lactobacillus plantarum P-8) | AGL65684.2 |
| Transposase (167aa, 98%, Lactobacillus paraplantarum) | CDF77674.1 | ||||||
| pBM2 | |||||||
| orf1 | 588 | 1 | 40.48 | 195 | 22538.9 | Putative integrase/recombinase (195aa, 99%, Lactobacillus casei) | WP_003586668.1 |
| Integrase (195aa, 96%, Lactobacillus sakei KCA311) | AJQ16980.1 | ||||||
| orf2 | 667 | 930 | 37.88 | 87 | 9981.6 | AbrB family transcriptional regulator (plasmid) (87aa, 100%, Lactobacillus plantarum HFC8) | ALG27235.1 |
| orf3 | 930 | 1277 | 39.66 | 115 | 13031.8 | PemK family protein (115aa, 96%, Lactobacillus plantarum HFC8) | ALG27234.1 |
| orf4 | 2063 | 1695 | 41.46 | 122 | 12265.3 | Prophage Lp1 protein 6 (122aa, 100%, Lactobacillus pentosus MP-10) | CCB84218.1 |
| Membrane protein (122aa, 99% , Lactobacillus plantarum) | WP_011101080.1 | ||||||
| orf5 | 2397 | 2143 | 31.76 | 84 | 10054.2 | Hypothetical protein (84aa, 99%, Lactobacillus plantarum) | WP_016527215.1 |
| orf6 | 2871 | 2407 | 37.63 | 154 | 17395.3 | Hypothetical protein (263aa, 98%, Lactobacillus plantarum) | WP_024002855.1 |
| orf7 | 3350 | 3853 | 46.63 | 167 | 18712.4 | Hypothetical protein (167aa, 99%, Lactobacillus plantarum subsp. plantarum P-8) | AGL65684.2 |
| Transposase (167aa, 98%, Lactobacillus paraplantarum) | CDF77674.1 | ||||||
| repB | 4829 | 4314 | 35.08 | 171 | 19956.9 | Replication protein RepB (171aa, 84%, Lactobacillus brevis) | WP_041816333.1 |
| Replication protein RepB (171aa, 84%, Lactobacillus brevis KB290) | BAN08207.1 | ||||||
| Replication protein (192aa, 66%, Pediococcus acidilactici H plasmid pSMB74) | AAP55633.1 | ||||||
| repA | 5814 | 4822 | 34.74 | 330 | 38652.7 | Initiator Replication family protein (311aa, 99%, Pediococcus pentosaceus) | WP_002834578.1 |
| Probable replication protein rep (plasmid) (376aa, 99%, Lactobacillus brevis KB290) | BAN08206.1 | ||||||
| Rep3 (plasmid pSMB74) (319aa, 82%, Pediococcus acidilactici) | CAA53278.1 | ||||||
| Replication protein A (plasmid pMD5057) (311aa, 81%, Lactobacillus plantarum 5057) | AAP55632.1 | ||||||
| orf10 | 6688 | 6476 | 32.86 | 70 | 8231.2 | Hypothetical protein (70aa, 97%, Lactobacillus nodensisi) | WP_025025371.1 |
| Hypothetical protein (70aa, 97%, Lactobacillus versmoldensis) | WP_010625611.1 | ||||||
| pBM3 | |||||||
| orf1 | 766 | 554 | 32.8 | 70 | 8369.7 | Hypothetical protein (70aa, 99%, Lactobacillus brevis) | WP_042253900.1 |
| Hypothetical protein (70aa, 96%, Lactobacillus nodensis) | WP_025025371.1 | ||||||
| orf2 | 1522 | 1163 | 36.1 | 119 | 14162.1 | Hypothetical protein (119aa, 99%, Lactobacillus plantarum) | WP_027821987.1 |
| Hypothetical protein (plasmid) (121aa, 99%, Lactobacillus brevis BSO 464) | AJA81476.1 | ||||||
| orf3 | 2122 | 1535 | 41 | 195 | 22475.8 | Integrase (195aa, 91%, Lactobacillus pentosus IG1) | CCC15451.1 |
| Integrase (195aa, 99%, Lactobacillus brevis) | WP_011669005.1 | ||||||
| orf4 | 2213 | 2494 | 36.5 | 93 | 10822.1 | Antitoxin (93aa, 100% , Lactobacillus brevis) | WP_011669006.1 |
| Antitoxin (pMK06) (92aa, 96%, Lactobacillus plantarum HFC8) | ALG27537.1 | ||||||
| orf5 | 2491 | 2847 | 37.5 | 118 | 13861.9 | Plasmid stabilisation system protein (118aa, 100% , Lactobacillus brevis) | WP_011669007.1 |
| Addiction module toxin RelE (pMK06) ( 118aa, 97%, Lactobacillus plantarum HFC8) | ALG27529.1 | ||||||
| orf6 | 2923 | 3300 | 36.7 | 125 | 14714.5 | Transposase, partial (112aa, 93% , Lactobacillus plantarum) | WP_046041026.1 |
| orf7 | 5296 | 3602 | 31.5 | 564 | 65233.9 | Cell surface protein (564aa, 99%, Lactobacillus plantarum) | WP_016527444.1 |
| orf8 | 5812 | 6300 | 37.8 | 162 | 18792 | Hypothetical protein (162aa, 95% , Lactobacillus plantarum) | WP_016511466.1 |
| Hypothetical protein (162aa, 94%, Lactobacillus rhamnosus) | WP_024306043.1 | ||||||
| orf9 | 7005 | 7385 | 37.3 | 126 | 14408.6 | Hypothetical protein (125aa, 97%, Lactobacillus plantarum) | WP_003646093.1 |
| Hypothetical protein (125aa, 86%, Lactobacillus plantarumWJL) | ERO39647.1 | ||||||
| orf10 | 7848 | 7345 | 46.8 | 167 | 18744.6 | Transposase (167aa, 99%, Lactobacillus plantarum subsp. plantarum P-8) | AGL65684.2 |
| Transposase (167aa, 99%, Lactobacillus plantarum) | WP_024272162.1 | ||||||
| pBM4 | |||||||
| orf1 | 662 | 129 | 39.89 | 177 | 20171.7 | Transposase (177aa, 99%, Lactobacillus plantarum) | WP_021731278.1 |
| Transposase (177aa, 91%, Lactobacillus paracasei) | WP_016385682.1 | ||||||
| orf2 | 1936 | 1106 | 28.28 | 276 | 32479.2 | Endonuclease (278 aa, 69%, Lactobacillus brevis) | WP_042749360.1 |
| orf3 | 2456 | 1998 | 28.1 | 152 | 17600.8 | Hypothetical protein (152aa, 68%, Lactobacillus plantarum) | WP_046947582.1 |
| orf4 | 4043 | 3540 | 38.89 | 167 | 19284.3 | Hypothetical protein (167aa, 98%, Lactobacillus plantarum) | WP_011031951.1 |
| Hypothetical protein (167aa, 91%, Lactobacillus plantarum) | WP_045353452.1 | ||||||
| orf5 | 4867 | 4400 | 43.59 | 155 | 18007.2 | Ferritin (155aa,100%, Lactobacillus brevis) | WP_042253884.1 |
| Stress induced DNA binding protein (155aa, 99%, Lactobacillus sakei KCA311) | AJQ16931.1 | ||||||
| orf6 | 6633 | 5422 | 28.22 | 403 | 46440.9 | Hypothetical protein (401aa, 75%, Lactobacillus florum) | WP_009166650.1 |
| orf7 | 7097 | 6795 | 33.33 | 100 | 11858.1 | Translation repressor RelE (100aa, 97%, Lactobacillus plantarum) | WP_021357787.1 |
| Translation repressor RelE (100aa, 99%, Lactobacillus versmoldensis) | WP_010625626.1 | ||||||
| orf8 | 7365 | 7087 | 36.92 | 92 | 10106 | RelB (92aa, 98%, Lactobacillus plantarum) | WP_027822890.1 |
| orf9 | 7469 | 8056 | 38.44 | 195 | 22447 | Integrase (195aa, 99%, Lactobacillus brevis) | WP_042253402.1 |
| Integrase (195aa, 96%, Lactobacillus pentosus MP-10) | CCB81910.1 | ||||||
| orf10 | 8480 | 8692 | 31.92 | 70 | 8273.3 | Hypothetical protein (70aa, 96%, Lactobacillus nodensis) | WP_025025371.1 |
| Hypothetical protein (70aa, 96%, Lactobacillus brevis) | WP_042253900.1 | ||||||
4.5. Relative Copy Number of pBM1-4
In this study, the relative copy numbers of pBM1, pBM2, pBM3, and pBM4 were determined by Droplet Digital PCR. Our results revealed that the relative copy numbers of pBM1, pBM2, pBM3, and pBM4 were approximately 82, 24, 34, and 16 copies per chromosome equivalent.
5. Discussion
In this work, four native plasmids were isolated from L. plantarum BM4. The sequence analysis results revealed that pBM1, pBM2, pBM3, and pBM4 were 6069 bp, 7042 bp, 8131 bp and 8891 bp in length, and we predicted 8 ORFs on pBM1, and 10 ORFs on pBM2, pBM3, and pBM4. According to our results, only pBM2 encoded an overlapping plasmid probable replication protein (repB) and an initiator replication family protein (repA). The repA protein (GenBank: ALO75838.1) of pMB2 shares 99% identity with the rep protein of plasmid pKB290-8 from Lactobacillus brevis KB290 (20), 80% identity with the repA protein of plasmid pUCL287 from Pediococcus halophilus ATCC33315 (21), 86% identity with the repA protein of plasmid pMD5057 from Lactobacillus plantarum 5057 (22), 78% identity with the repA protein of plasmid pRV500 from L. sakei, 81% identity with the repB protien of plasmid pSMB74 from Pediococcus acidilactici H (23), and 78% identity with the initiation protein repA of plasmid pRS5 from P. pentosaceus (24), all of which belong to the theta-replicating group.
Moreover, a typical ori was predicted upstream of repA (sites: 5749 - 6000), which consists of three direct 11-bp repeats (three times) and four 22-bp iterons, tandemly repeated (Figure 4). It has been reported that the 22-bp introns are important for pSB01 and the 11-bp repeats are necessary for pUCL287 replication (Benachour et al. 1997) (14, 25). Interestingly, the overlap of genes coding repA and repB was found in several theta-replicating plasmids, such as pKB290-8 from Lactobacillus brevis KB290, plasmid pSMB74 from Pediococcus acidilactici H, and plasmid pUCL287 from Pediococcus halophilus ATCC33315. It has been previously reported that RepB in pUCL287 is involved in plasmid stability and regulation of plasmid copy number, but is not essential for replication. In conclusion, these features suggest that pBM2 belongs to the group of pUCL287 theta-replicating plasmids (13, 26).
All four plasmids contain a similar INT_C_like_3 Integrase protein of 195aa, which has been previously reported that integrase in pSMB 74 is likely to be involved in recombination or integration (23). Tyrosine recombinase (integrase) belongs to a DNA breaking-rejoining enzyme superfamily. Many DNA breaking-rejoining enzymes also have N-terminal domains, which show little sequence or structure similarity (27). A toxin-antitoxin (TA) plasmid maintenance system was identified in pBM1 and pBM3, which belong to plasmid stabilization system protein in gram-negative and gram-positive bacteria, and may play a role in keeping low copy-number plasmids stable through neutralization toxin (2). The exact molecular function of these proteins is not known. This family also encompasses RelE/ParE, which seems to occur in pairs and to be organized as an operon (28).
Orf2-encoding protein in pBM2 belongs to antidote-toxin recognition MazE, which is the antidote to the toxin MazF of E. coli, and regulates the prokaryotic chromosomal addiction module. MazE-MazF is thought to play a role in programmed cell death when cells suffer nutrient deprivation, and MazE-MazF modules have been implicated in the bacteriostatic effects of other addiction modules (19). Orf3 in pBM2 encodes a PemK-like protein, which is an inhibitor for growing in E. coli known to bind to the promoter region of the Pem operon, auto-regulating synthesis. This Pfam family consists of the PemK protein in addition to ChpA, ChpB, and other PemK-like proteins (19).
Our data do not support the replication mechanism of plasmids pBM1, pBM3, or pBM4. However, it was reported that some plasmids without replication protein replicate via an RNA-based replication mechanism, such as pCD033 from L. plantarum 3NSH, and plasmid pColE1 (29-31). The principle of ddPCR involves subdividing a single PCR reaction mixture into many small partitions and each undergoing the PCR reaction separately (32). Several studies have reported using ddPCR to determine the relative copy numbers of plasmids (33, 34). According to our results, the relative copy numbers of pBM1, pBM2, pBM3, and pBM4 were calculated as 82, 24, 34, and 16 copies per chromosome equivalent using the ddPCR method. Our results suggest that pBM1-4 belong to the low copy plasmids.
6. Conclusion
In summary, we characterized four new plasmids, pBM1, pBM2, pBM3, and pBM4, isolated from L. plantarum BM4. By sequence analysis and comparison, we found that, only pBM2 contained replication protein RepB and rep3, which coding for a putative initiator protein contained a putative 11-and 22-bp repeat origin of replication segment, indicating the pBM2 belongs to the pUCL287 subfamily of theta-type replicons. Moreover, other proteins in pBM1-4 also involved module toxin, antitoxin, integrase, AbrB family transcriptional regulator, transposase, endonuclease, and translation repressor RelE. The relative copy numbers of pBM1-4 were estimated to be 82, 24, 34, and 16 copies, respectively.
References
-
1.
Klarin B, Molin G, Jeppsson B, Larsson A. Use of the probiotic Lactobacillus plantarum 299 to reduce pathogenic bacteria in the oropharynx of intubated patients: a randomised controlled open pilot study. Crit Care. 2008;12(6):R136. [PubMed ID: 18990201]. https://doi.org/10.1186/cc7109.
-
2.
Siezen RJ, Francke C, Renckens B, Boekhorst J, Wels M, Kleerebezem M, et al. Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. J Bacteriol. 2012;194(1):195-6. [PubMed ID: 22156394]. https://doi.org/10.1128/JB.06275-11.
-
3.
Fan J, Xi X, Huang Y, Cui Z. Isolation of a minireplicon of the plasmid pG6303 of Lactobacillus plantarum G63 and characterization of the plasmid-encoded Rep replication protein. J Genet. 2015;94(2):177-86. [PubMed ID: 26174665].
-
4.
Zhou H, Hao Y, Xie Y, Yin S, Zhai Z, Han B. Characterization of a rolling-circle replication plasmid pXY3 from Lactobacillus plantarum XY3. Plasmid. 2010;64(1):36-40. [PubMed ID: 20353802]. https://doi.org/10.1016/j.plasmid.2010.03.003.
-
5.
Daming R, Yinyu W, Zilai W, Jun C, Hekui L, Jingye Z. Complete DNA sequence and analysis of two cryptic plasmids isolated from Lactobacillus plantarum. Plasmid. 2003;50(1):70-3. [PubMed ID: 12826059].
-
6.
de las Rivas B, Marcobal A, Muñoz R. Complete nucleotide sequence and structural organization of pPB1, a small Lactobacillus plantarum cryptic plasmid that originated by modular exchange. Plasmid. 2004;52(3):203-11. https://doi.org/10.1016/j.plasmid.2004.09.001.
-
7.
Eguchi T, Doi K, Nishiyama K, Ohmomo S, Ogata S. Characterization of a phage resistance plasmid, pLKS, of silage-making Lactobacillus plantarum NGRI0101. Biosci Biotechnol Biochem. 2000;64(4):751-6. [PubMed ID: 10830488]. https://doi.org/10.1271/bbb.64.751.
-
8.
Ma X, Li J, Xiong Y, Zhai Z, Ren F, Hao Y. Characterization of a Rolling-Circle Replication Plasmid pM411 from Lactobacillus plantarum 1-3. Curr Microbiol. 2016;73(6):820-6. [PubMed ID: 27592105]. https://doi.org/10.1007/s00284-016-1124-7.
-
9.
Ruiz-Maso JA, Lopez-Zumel C, Menendez M, Espinosa M, del Solar G. Structural features of the initiator of replication protein RepB encoded by the promiscuous plasmid pMV158. Biochim Biophys Acta. 2004;1696(1):113-9. [PubMed ID: 14726211].
-
10.
Sanchez C, Hernandez de Rojas A, Martinez B, Arguelles ME, Suarez JE, Rodriguez A, et al. Nucleotide sequence and analysis of pBL1, a bacteriocin-producing plasmid from Lactococcus lactis IPLA 972. Plasmid. 2000;44(3):239-49. [PubMed ID: 11078650]. https://doi.org/10.1006/plas.2000.1482.
-
11.
Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53(2):126-36. [PubMed ID: 15737400]. https://doi.org/10.1016/j.plasmid.2004.12.008.
-
12.
van Belkum MJ, Stiles ME. Characterization of the theta-type plasmid pCD3.4 from Carnobacterium divergens, and modulation of its host range by RepA mutation. Microbiology. 2006;152(Pt 1):171-8. [PubMed ID: 16385127]. https://doi.org/10.1099/mic.0.28294-0.
-
13.
Alpert CA, Crutz-Le Coq AM, Malleret C, Zagorec M. Characterization of a theta-type plasmid from Lactobacillus sakei: a potential basis for low-copy-number vectors in lactobacilli. Appl Environ Microbiol. 2003;69(9):5574-84. [PubMed ID: 12957947].
-
14.
Benachour A, Frere J, Flahaut S, Novel G, Auffray Y. Molecular analysis of the replication region of the theta-replicating plasmid pUCL287 from Tetragenococcus (Pediococcus) halophilus ATCC33315. Mol Gen Genet. 1997;255(5):504-13. [PubMed ID: 9294035].
-
15.
Bruand C, Le Chatelier E, Ehrlich SD, Janniere L. A fourth class of theta-replicating plasmids: the pAM beta 1 family from gram-positive bacteria. Proc Natl Acad Sci U S A. 1993;90(24):11668-72. [PubMed ID: 8265606].
-
16.
Meijer WJ, de Boer AJ, van Tongeren S, Venema G, Bron S. Characterization of the replication region of the Bacillus subtilis plasmid pLS20: a novel type of replicon. Nucleic Acids Res. 1995;23(16):3214-23. [PubMed ID: 7667098].
-
17.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9. [PubMed ID: 17488738]. https://doi.org/10.1093/molbev/msm092.
-
18.
Zhang H, Hao Y, Zhang D, Luo Y. Characterization of the cryptic plasmid pTXW from Lactobacillus paracasei TXW. Plasmid. 2011;65(1):1-7. [PubMed ID: 20709099]. https://doi.org/10.1016/j.plasmid.2010.08.002.
-
19.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, et al. CDD: NCBI's conserved domain database. Nucleic Acids Research. 2014;43(D1):D222-6. https://doi.org/10.1093/nar/gku1221.
-
20.
Fukao M, Oshima K, Morita H, Toh H, Suda W, Kim SW, et al. Genomic analysis by deep sequencing of the probiotic Lactobacillus brevis KB290 harboring nine plasmids reveals genomic stability. PLoS One. 2013;8(3). e60521. [PubMed ID: 23544154]. https://doi.org/10.1371/journal.pone.0060521.
-
21.
Benachour A, Frere J, Boutibonnes P, Auffray Y. Characterization and replication mode determination of the minimal replicon of Tetragenococcus halophila ATCC33315 plasmid pUCL287. Biochimie. 1995;77(11):868-74. [PubMed ID: 8824766].
-
22.
Danielsen M. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid. 2002;48(2):98-103. [PubMed ID: 12383727].
-
23.
Motlagh A, Bukhtiyarova M, Ray B. Complete nucleotide sequence of pSMB 74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett Appl Microbiol. 1994;18(6):305-12. [PubMed ID: 7764941].
-
24.
Teresa Alegre M, Rodriguez MC, Mesas JM. Characterization of pRS5: a theta-type plasmid found in a strain of Pediococcus pentosaceus isolated from wine that can be used to generate cloning vectors for lactic acid bacteria. Plasmid. 2009;61(2):130-4. [PubMed ID: 19027788]. https://doi.org/10.1016/j.plasmid.2008.10.002.
-
25.
Nakamura M, Ogata K, Nagamine T, Tajima K, Matsui H, Benno Y. The replicon of the cryptic Plasmid pSBO1 isolated from Streptococcus bovis JB1. Curr Microbiol. 2001;43(1):11-6. [PubMed ID: 11375657]. https://doi.org/10.1007/s002840010252.
-
26.
Crutz-Le Coq AM, Zagorec M. Vectors for Lactobacilli and other Gram-positive bacteria based on the minimal replicon of pRV500 from Lactobacillus sakei. Plasmid. 2008;60(3):212-20. [PubMed ID: 18789962]. https://doi.org/10.1016/j.plasmid.2008.08.002.
-
27.
Van Houdt R, Leplae R, Lima-Mendez G, Mergeay M, Toussaint A. Towards a more accurate annotation of tyrosine-based site-specific recombinases in bacterial genomes. Mob DNA. 2012;3(1):6. [PubMed ID: 22502997]. https://doi.org/10.1186/1759-8753-3-6.
-
28.
Sorvig E, Skaugen M, Naterstad K, Eijsink VG, Axelsson L. Plasmid p256 from Lactobacillus plantarum represents a new type of replicon in lactic acid bacteria, and contains a toxin-antitoxin-like plasmid maintenance system. Microbiology. 2005;151(Pt 2):421-31. [PubMed ID: 15699191]. https://doi.org/10.1099/mic.0.27389-0.
-
29.
Heiss S, Grabherr R, Heinl S. Characterization of the Lactobacillus plantarum plasmid pCD033 and generation of the plasmid free strain L. plantarum 3NSH. Plasmid. 2015;81:9-20. [PubMed ID: 26038184]. https://doi.org/10.1016/j.plasmid.2015.05.004.
-
30.
Jahn M, Vorpahl C, Hubschmann T, Harms H, Muller S. Copy number variability of expression plasmids determined by cell sorting and Droplet Digital PCR. Microb Cell Fact. 2016;15(1):211. [PubMed ID: 27993152]. https://doi.org/10.1186/s12934-016-0610-8.
-
31.
Tomizawa J, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981;78(10):6096-100. [PubMed ID: 6171811].
-
32.
Brantl S. Plasmid Replication Control by Antisense RNAs. Microbiol Spectr. 2014;2(4):PLAS-1-2013. [PubMed ID: 26104196]. https://doi.org/10.1128/microbiolspec.PLAS-0001-2013.
-
33.
Dong L, Meng Y, Sui Z, Wang J, Wu L, Fu B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci Rep. 2015;5:13174. [PubMed ID: 26302947]. https://doi.org/10.1038/srep13174.
-
34.
Jahn M, Vorpahl C, Turkowsky D, Lindmeyer M, Buhler B, Harms H, et al. Accurate determination of plasmid copy number of flow-sorted cells using droplet digital PCR. Anal Chem. 2014;86(12):5969-76. [PubMed ID: 24842041]. https://doi.org/10.1021/ac501118v.