Abstract
Background:
Methicillin-resistant Staphylococcus aureus (MRSA) not only is a notorious pathogen in clinical settings but also is an environmental issue that its presence in environmental wastewater is highlighted by several reports. Due to the negative impacts of antibiotics, alternatives like bacteriophages, as biocontrol, are considered safe. However, not all bacteriophages are safe. Thus, the characterization of bacteriophages is necessary.Objectives:
This study aimed to, firstly isolate MRSA from wastewater and, secondly to perform bacteriophage isolation from the water samples to investigate its physical and genomic characteristics.Methods:
Water samples were collected from seven locations across Nagpur city, India, bacteria were isolated on the S. aureus specific agar. For detecting MRSA, we followed the disc diffusion method. Isolation of bacteriophage against MRSA was performed by a modified enrichment method. We investigated its physical characteristics by the one-step growth rate, adsorption rate, host range, survivability, electron microscopy, and genomic sequencing for bioinformatics analysis.Results:
Four MRSA were isolated from wastewater samples. We got a bacteriophage against an MRSA from the river Ganga. The bacteriophage belongs to the Podoviridae family, subfamily Autographivirinae. It was stable till 40°C and could survive at a highly alkaline pH. It is specific to its host. The bacteriophage DNA encodes 52 ORF, and all predicted genes are on the same strand; it also encodes a phage RNA polymerase.Conclusions:
It is the first report of an S. aureus bacteriophage that belongs to the sub-family Autographivirinae. Our study and literature survey conclude that S. aureus bacteriophages of the Podoviridae family are safe for various downstream applications.Keywords
Bacteriophage Isolation Bacteriophage Methicillin-resistant, Staphylococcus aureus
1. Background
Staphylococcus aureus is one of the most successful pathogens due to its vast arsenal of virulence factors, including anchor proteins, secreted toxins and enzymes, polysaccharides, and immune system modulators (1). Methicillin, a semi-synthetic β-lactam antibiotic, was first applied to treat S. aureus infections, but the bacterium soon developed resistance and became known as methicillin-resistant Staphylococcus aureus (MRSA) (1), which not only is a notorious pathogen in clinical settings and the food industry but also is an environmental issue. Several reports have confirmed the presence of MRSA in wastewater and reported it as a serious environmental issue (2-5). Wastewater treatment plants (WWTPs) are considered hotspots of multi-drug resistant (MDR) bacteria (5). Irrational use of antibiotics is not only harmful to our health but also to the environment. There is a broad spectrum of antibiotics that can negatively affect commensal bacteria too. Moreover, antibiotic residues in the wastewater cause selective pressure, which in turn results in the propagation of drug resistance genes and MDR bacteria (5).
In 2015, the World Health Organization (WHO) introduced an action plan against antimicrobial resistance, wherein one of its critical focal points is the development of novel antimicrobial products to combat multidrug-resistant strains (1). Bacteriophages / phages (viruses of bacteria) are good candidates for controlling pathogenic bacteria because they kill specific host bacteria and do not disturb commensal bacteria. Phages are of two types, namely virulent and lysogenic, based on the lifecycle (1). Virulent phages are obligatory lytic phages. However, lysogenic phage, also called temperate phage, integrates its genome in the bacterial chromosome and forms prophage; on the onset of unfavorable conditions, they disintegrate from the bacterial host genome. Lysogenic phages are involved in the propagation of harmful genes by the process of transduction. Thus, for a phage to be considered as biocontrol, only a lytic phage must be chosen.
Phages are present in all conceivable environments. Isolating phage against S. aureus is a daunting task, which its difficulty is highlighted by researchers (6, 7). Even on getting Staphylococcus phage, their plaque size is extremely small or turbid, making the process of isolation and purification of phage difficult (8). The reason can be the presence of predominant Enteric group phages in wastewater (8, 9). All S. aureus phages belong to the order Caudovirales (1); The phages of S. aureus have linear double-stranded DNA, and based on tail morphology, they are grouped into three families: Myoviridae, which has long contractile double sheath tail; Siphoviridae, which has long non-contractile tail and, Podoviridae, that has a small contractile tail (10).
Temperate phages of S. aureus belong to the Siphoviridae family and are not considered for their application. Phages of S. aureus that belong to the Myoviridae and Podoviridae families are mostly lytic. These are important for downstream applications because a highly potent lytic phage is desirable as it can subvert the essential metabolic pathways of the host (11). An updated and comprehensive study on the diversity of staphylococcal phages has been done by Oliveira et al.; they grouped the staphylococcal phages into four major clusters of A to D and several sub-clusters (12). For application as an antimicrobial, genome analysis is essential. A phage must not have resistance, virulence genes, and integrase genes.
2. Objectives
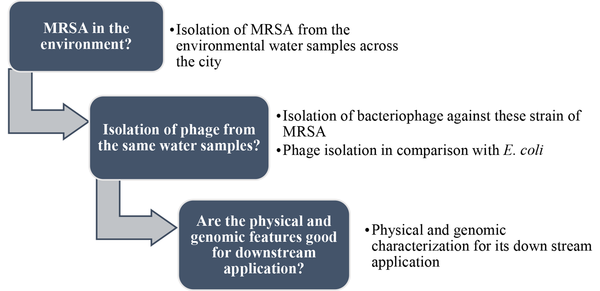
In this study, we isolated MRSA from wastewater samples and screened the wastewater samples for the presence of phages against the isolates. Upon difficulty in obtaining phage from the wastewater samples, we chose river water. Hence, we got the phage from the river sample; it was named vb_Sau_ARW1 (or ARW1) as per the guidelines of the International Committee on Taxonomy of Viruses (ICTV). Afterward, we investigated its physical and genomic characteristics. A flow diagram, Figure 1, is given to get an idea about this study.
A flow diagram depicting the work-plan of this research.
3. Methods
All water samples were collected through sterile practices. The experiments were done in triplicate. In graphical representations, the average value was taken. All culture media, chemicals, and ATCC bacterial strains were purchased from Himedia Labs Pvt. Ltd., Mumbai, India.
3.1. Isolation of Staphylococcus aureus
Wastewater and lake water samples were taken from seven locations across Nagpur city, India, to isolate S. aureus. Initially, 100 µL of a two-fold diluted sample was taken and spread plated on mannitol salt agar (MSA) and Vogel-Johnson agar (VJA). Yellow colour colonies on MSA plates and black-coloured colonies on VJA plates indicated putative S. aureus. Gram stain was used for identification, biochemical tests through the HiStaph identification kit. The kit consists of twelve wells with the reagent and agar; we added 50 µL of actively growing culture and kept the kits for overnight incubation. Then, we referred to the chart provided with the kit to identify the species of Staphylococcus by observing the colour change. We also did a multiplex PCR, MRSA semi-Q PCR kit (multiplex) (Himedia Labs Pvt. Ltd., Mumbai, India). Escherichia coli was also isolated through membrane filtration technique on MEC agar.
3.2. Resistance Profiling or the Antibiotic Susceptibility Test
Antibiotic susceptibility test (AST) of the confirmed S. aureus (that is, isolates W1-W4) with a standard strain S. aureus ATCC 25923 as control was done by Kirby Bauer disk diffusion method, as per the guidelines of Clinical Laboratory Standard Institute (CLSI) with the following antibiotics having concentrations in μg/disk: vancomycin (VA30), cefepime (CPM30), cefuroxime (CXM30), cefprozil (CPR30), gentamicin (GEN10), tobramycin (TOB30), erythromycin (E15), clindamycin (CD2), ciprofloxacin (CIP5), ceftriaxone / tazobactam (CIT30/10), tetracycline (TE30), chloramphenicol (C30), oxacillin (OX5), and ampicillin (AMP10) (Himedia Labs Pvt. Ltd., Mumbai, India).
3.3. 16S rRNA Sequencing was Done for the Isolate W1
The 6S rRNA gene was amplified by 27F and 1492R primers. A single discrete PCR amplicon band of 1500 bp was observed. Forward and reverse DNA sequencing reaction of PCR amplicon was carried out with forward primer and reverse primers using BDT v3.1 cycle sequencing kit on ABI 3730xl genetic analyzer (Eurofins Genomics, Bangalore, India).
3.4. Phage Isolation
We did the screening for phages in the water sample against the S. aureus isolates compared to Escherichia coli. The method proposed by Jensen et al. and the Enrichment method were slightly modified and followed (13, 14). Firstly, we filtered each sample through a sterile membrane filter of porosity 0.45 µm (Merck Millipore). In a 50 mL flask, we added 15 mL of the membrane filtered water sample, 7.5 mL of actively growing host culture (optical density 0.4 OD at wavelength 600 λ), and 7.5 mL of double-strength tryptic soy broth (TSB). It was kept overnight in a rotator incubator at 37°C and 60 rpm/min, followed by centrifugation at 6000 rpm at 4°C for 15 min. The suspension was filtered through a sterile 0.22 µm membrane filter. Then, 100 µL of this filtrate was mixed with 100 µL of the bacterial host and kept in a rotator incubator at 37°C before adding over the top agar of the tryptic soy agar (TSA) plate; this phage enumeration method (plaque assay) is called double agar overlay method (12, 15).
3.5. Host Range of the Phage ARW1
Host range is the ability of the phage to infect other bacterial hosts. For this, the following environmental S. aureus isolates were taken: W1, W2, W3, W4, E. coli ATCC 23848, S. aureus ATCC 25923, and a lab strain of Enterobacter. Afterward, 8 µL of 0.22 µm membrane filtered ARW1 lysate was added to the host culture plates. A clear zone on the culture plate might indicate a phage active against that host. Agar overlay was done to exclude the doubt of lysis due to lysozyme in the lysate. For this, ARW1 lysate was incubated with isolates mentioned above and kept for overnight incubation at 37°C in a rotator incubator, and agar overlay was done the next day. In case of ambiguity, such as a faint or turbid plaque, further analysis by picking the plaque and re-incubating it with the host, followed by enumeration by double agar overlay.
3.6. Partial Purification of ARW1
Phage purification is done by picking a single plaque and re-enriching with the host three consecutive times and precipitating by PEG-NaCl for downstream applications. For partial purification and concentration of phages (16), ten plates (agar overlay plating method) with dense plaque count were used. In each plate, 4 mL Phage buffer (P buffer) (10 Mm Tris-Cl, 10 mM MgSO4, 68 mM NaCl, 1 mM CaCl2 constituted in molecular grade water and made up the volume to 1 L) was added and kept overnight at 4°C. The lysate was centrifuged at 6000 × g for 15 min at 4°C and was filtered through a 0.22 µm membrane filter. In the filtrate, RNase A (20 mg/mL) and DNase (1 mg/mL) were added such that the final concentration was 4 µL/mL. The filtrate was kept at 37°C for 30 minutes and then was maintained at room temperature for an additional one hour. Further, for phage precipitation, an ice-cold solution of 40% PEG-NaCl was added (PEG-6000 and 3M NaCl) such that its volume was one-fourth of the total volume of the filtered lysate. This was kept at 4°C on ice for an hour, followed by centrifugation at 6000 × g for 40 min at 4°C. The precipitate was reconstituted in 0.1 mL of P buffer. This partially purified phage was kept at -80°C for the next phase of analysis as described below.
3.7. Transmission Electron Microscopy
We did transmission electron microscopy analysis at the Indian Institute of Technology, Roorkee, India. Sample prepared briefly as -10 µL (1x108 PFUs/mL) of the PEG precipitated phage and purified crude lysate was loaded on a copper grid, allowed it to air-dry and stained with 10 µL of uranyl acetate (17). Imaging was performed at magnification 35000X for crude lysate and at 15000X for PEG precipitated samples, at an operating voltage of 120kV (TEM model: Tecnai G2 20).
3.8. ARW1 Genomic DNA Isolation and Sequencing
The genomic DNA was extracted through phenol-chloroform extraction and ethanol precipitation method (16). The precipitate was reconstituted in molecular-grade water and stored at -20°C. The quality of the genomic DNA was evaluated on 0.8% agarose gel (loaded three μL) for the single intact band. The gel was run at 110 V for 30 min. Phage genomic sequencing and analysis of high-quality data (HQ) was done at the Xcelris Labs Pvt. Ltd., India. It was sequenced on the Illumina platform with a read length 2X 150 PE and coverage of more than 10000. For de novo assembly of the phage, SPAdes version–3.1.0 was used. Similarity search through BLASTn and BLASTx (version 2.2.28) was done. Genes were predicted through Prodigal version 2.6.3 (18). The phage lifecycle was predicted through a web-based tool, PHACTS (19). The presence / absence of tRNA genes was ascertained through a web-based tool called tRNAscan-SE (17, 20). A web-based tool was used to identify antimicrobial resistance genes Resistance Gene Identifier (RGI) (21).
3.9. Adsorption Rate
To determine adsorption rate (8, 22), 9 mL of exponentially growing S. aureus (W1) culture of optical density at 600 nm (OD600) 0.2 OD (108 CFUs/mL) was taken in a 100 mL flask, to this 1 mL of pre-warmed phage suspension of titre 1 × 105 PFUs/mL was added. At time zero, 100 µL aliquot was withdrawn and added to 900 µL fresh broth (kept in 2 mL Eppendorf tube), thereafter at regular intervals of 1.5 min, this process was repeated till 10 min. These aliquots were centrifuged at 16000 × g rpm for 10 min. The supernatant was carefully withdrawn and mixed with fresh host broth culture for phage enumeration. The amount of phage adsorbed to the host cell was determined as a decrease in the titre of free phage. Refer to Appendix 1 in Supplementary File. Adsorption rate (ka) was calculated with the mathematical formula of
Which B is host concentration, t is the time for decreased plaque counts from initial, Po to the final plaque count, P.
3.10. One-step Growth
For one-step growth analysis (23, 24), 1 mL of phage at an MOI of 0.1 was added in 9 mL of exponentially growing S. aureus culture (similar to adsorption rate method) and allowed to adsorb completely on S. aureus for 8 min; followed by centrifugation at 6000 × g for 10 min. This pellet was suspended in 10 mL broth. After every 5 minutes, 100 µL of the aliquot was taken and added in 900 µL broth and plated by agar overlay method. An average value of five independent experiments was taken.
3.11. Phage Viability
The one-year viability of ARW1 was evaluated. Filtered phage stocks were kept in the P buffer. Their viability was tested at temperatures ranging from -20°C, 4°C, 25°C, 40°C, and 60°C. Survivability of phage was studied at different pH: in the 9 mL of P buffer of different pH was prepared, that is, pH of 2, 4, 6, 8, 10, and 12, to this 1 mL of filtered phage lysate was added and kept at 37°C for overnight incubation. Post incubation, 100 µL of the suspension was taken from each tube and mixed with 100 µL of actively growing host culture, and agar-overlay was done for phage enumeration.
4. Results
4.1. Isolation of Staphylococcus aureus and Its Phage from Water Samples
For the geographical coordinates of sampling sites, bacterial host identification, and AST, refer to Appendix 2 in Supplementary File, were performed. The results of biochemical and PCR tests are given in Appendices 3 and 4 in Supplementary File. We confirmed four S. aureus isolates through biochemical tests, and for one isolate (against which we got a phage), 16S rRNA sequencing was performed, and the sequence was deposited at the GenBank (submission ID- MN078268). These four isolates were found to be resistant to oxacillin; If an isolate is resistant to oxacillin means, it is resistant to methicillin. Thus, through AST, we ascertained them to be methicillin-resistant.
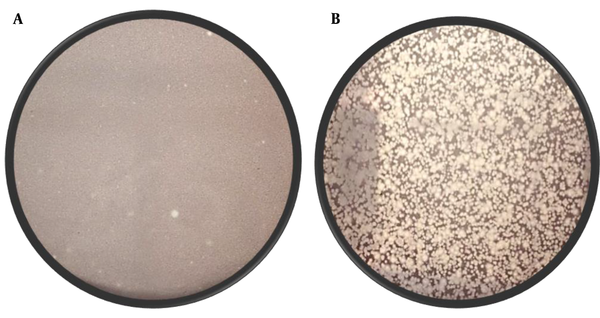
We compared the screening of phages from the water samples for S. aureus isolates to E. coli (with eleven E. coli isolates). No lytic phage against S. aureus was found in the screening process, but a few lytic phages were obtained against E. coli (from the city water samples). The results of the phage screening from the water samples are summarised in Supplementary File (Appendices 5 and 6). However, from the River Ganga, we got phage against S. aureus (W1 isolate) with plaque count 38PFU/mL, and the density of phage against E. coli ATCC 23848 was too numerous to count (TNTC). In Figure 2, representative plates are given to show the difference in isolating the phages of S. aureus (W1 isolate) and E. coli.
The presence of clear spots (plaque) over the lawn of bacteria confirms the phage. The image on the left is a representative plate of phage got from the River Ganga against the MRSA (arrow showed at the zone of clearing is a plaque); The plaques against MRSA are small and faint. The image on the right is a representative plate of phage got from the River Ganga against E. Coli ATCC 23848; The plaques against this bacterium are numerous and have clear morphology.
4.2. Host Range
We observed clear plaques for the entire duration of this study against the S. aureus (W1 isolate), but faint plaques were observed with the other three S. aureus isolates as well as S. aureus ATCC 25923. But no plaques were observed against E. coli ATCC 23848 and Enterobacter. It was neither infective against any of the 11 E. coli isolates. Upon subsequent plating, the plaques with faint morphology yielded no plaque. The phage could only infect the W1 isolate.
4.3. Identification Through TEM
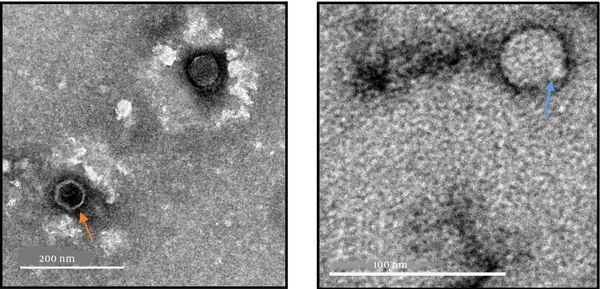
The TEM images of the phage, at different scales and magnifications, are shown in Figure 3, which shows that the phage belongs to the order Caudovirales and Podoviridae family. It has a typical icosahedral geometry and a short stubby tail.
Image at the left panel of the phage ARW1 from PEG precipitated crude lysate at a magnification 15000X; Orange arrow indicates the phage particle devoid of its genome as the inside is hollow. The image on the right panel is of crude lysate at a magnification of 35000X shows the phage with a stubby tail and icosahedral head (arrow shows phage particle).
4.4. Identification of ARW1
Through de novo assembly, we got the largest Scaffold of 44323 bp (51.43% G + C content). We did BLAST to search for similarity, which revealed a strong resemblance to a group of recently identified phages belonging to the Podoviridae family, Autographivirinae sub-family. The Phage ARW1 genomic DNA sequencing data is deposited at the GenBank, SRA submission ID: PRJNA637459.
4.5. Gene Predictions / ORFs
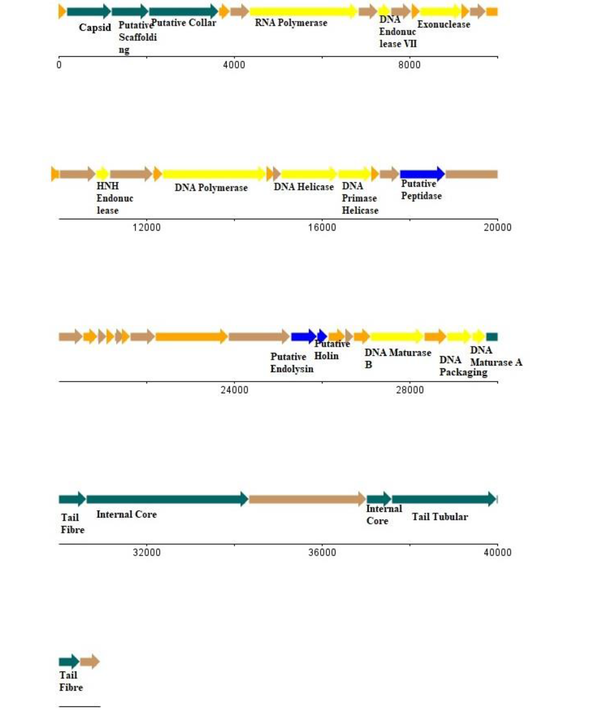
Through the Prodigal tool, we predicted that 52 ORFs can be grouped into-phage structural module, lysis system, replication-transcription module, as well as several hypothetical genes. Of note, all ORFs are presented on one strand, which is (–) strand; it also encodes an RNA polymerase. The phage genome does not have any tRNA gene. Also, AMR genes were absent. All predicted genes were plotted using the DNA plotter (25), refer to Figure 4. The phage lifecycle prediction tool PHACTS non-confidently classified it as a lytic phage.
A linear map of vb_Sau_ARW1 genome. Genes were predicted through Prodigal (Plotted on the software DNA Plotter).
4.6. Physical Parameters
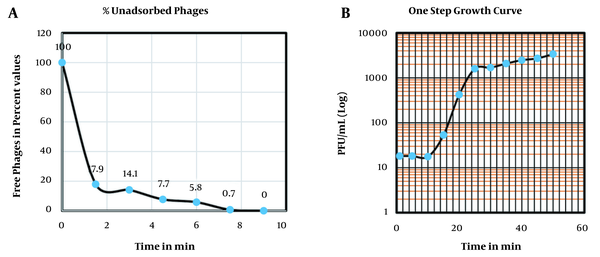
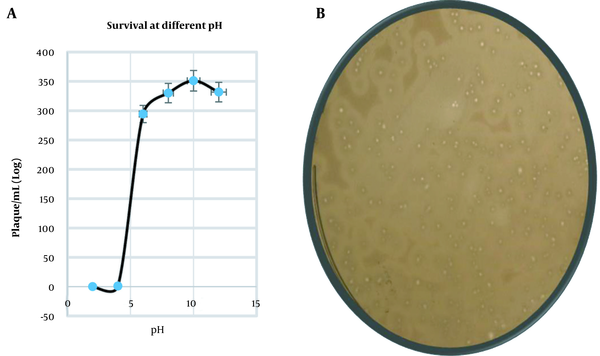
From the adsorption assay, the adsorption rate was 6.72 × 10-9 mL/min. As shown in Figure 5A, it can be estimated that in 7 minutes, 99% of phage was adsorbed; Thus, we gave 7 minutes for the phage and the host to adsorb before the one-step growth curve, growth curve followed the usual sigmoidal trend. The latency period was found to be 12 minutes, with an average burst size of 200 PFU/mL (Figure 5B). Phage survival was accessed at different conditions. Phage was viable in the deep freeze for the entire duration of this study for three years when kept at -20°C or -80°C and remained active for one month when held at 4°C. Its titre was unchanged for one month at 4°C. Activity reduced when it was kept at 60°C for 10 min. It was estimated that a decrease of 70.9% occurred compared to the average phage titre taken at room temperature. Phage survival at different pH (ie, 2, 4, 6, 8, 10, and 12) was done and is active at alkaline pH (Figure 6A), a representative plate of a decrease in the plaque morphology at an acidic pH is shown in Figure 6B.
(A) Graph of adsorption assay of the phage determined in terms of percent free phages; (B) One-step growth curve of the phage ARW1.
(A) Graph representing the phage survival at different acidity and basicity; (B) A plaque assay plate shows a considerable decrease in the plaque size in the acidic medium.
5. Discussion
Our findings corroborate the earlier research that reported challenges regarding finding a phage against S. aureus; It is an exciting phenomenon to delve deeper into. We may hypothesize that isolating phage against the already low concentrated host is a daunting task, as we know that the phage exists where its host exists. In this case, although MRSA has been isolated from environmental wastewater, it was in a low concentration and challenging to grow. On the other hand, many S. aureus strains often have many prophages in their genomes, which makes the entry of new phages difficult. The River Ganga holds a special position in the history of phages (26). Our lab has isolated phage against Escherichia spp. from the ‘Gomukh’-the origin point of the River Ganga- (the melting Himalayan Permafrost), so it is considered the most critical reservoir of phage (26). Therefore, the River Ganga water sample was selected for phage isolation.
As seen in Figure 2, phages against E. coli are abundant, but phages against S. aureus are faint and scanty; purifying the ARW1 phage was also a difficult task. Also, several morphological types of plaques are present in both plates, indicating that various phages may be presented against the same host. This difference in finding phage can also be attributed to the organismal composition of man-made sewage systems and a natural river system. The former is richer in enteric groups (27), whereas a natural river system is richer in species compositions. Both genomic and TEM data are in accordance, ie, ARW1 has characteristics of order Caudovirales, family Podoviridae. Like a typical phage, it has a modular genome organization (Figure 4). The genes are present in a particular order in which their transcription and replication are necessary, except for one putative holin gene. Usually, holin is present before endolysin; but it appears to be after endolysin; which is an exceptional feature. Additionally, this phage has an ORF for endopeptidase.
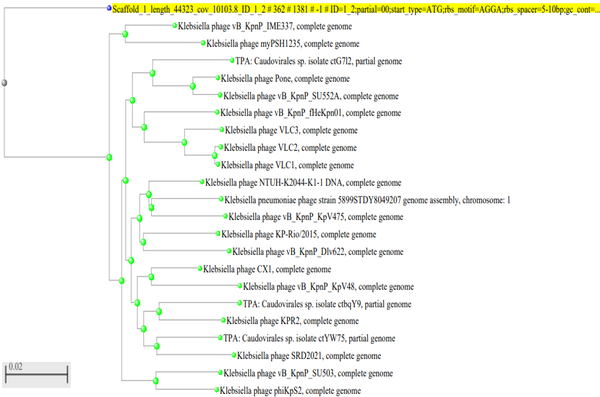
ARW1 genome has an RNA polymerase, which is an essential feature of the sub-family Autographivirinae. All predicted genes are encoded on the minus strand; This is a typical feature of the sub-family Autographivirinae. A few other phages of this sub-family are T7 and SP6 (28), a few phages belong to this group. Until now, no known phage of S. aureus belonging to this group has been recognized. Moreover, very few S. aureus phages of the family Podoviridae are known; Only 7% of Podoviridae phages of Staphylococcus (including other species of this genus) are known (29). S. aureusPodoviridae phages are an asset as they have small genomes that are amenable to genetic manipulations. Until now, no Podoviridae phages of S. aureus have any virulence or toxin genes, and they are strictly lytic, a desirable feature for downstream application (29). This phage forms a different branch on phylogenetic analysis of the phage capsid gene (phylogenetic tree based on BLASTn search and with default parameters). It is separate from the remaining phage groups. Thus, this seems to have diverged early in evolution, refer to Figure 7.
Phylogenetic tree constructed with the default parameters after BLASTn of the capsid protein gene. The phage as represented as scaffold_1 (highlighted), forms a distinct branch.
All phages belonging to Autographivirinae sub-family are of Gram-negative bacteria. All phages of Gram-negative bacteria have an additional lytic enzyme in their genomes called spanin; This is the first enzyme that acts upon the outer membrane of the Gram-negative bacterium (30). Interestingly, our phage genome does not have ORF for spanin (as the Gram-positive bacteria do not have an outer membrane). Recently, a Jumbo phage (their genomes are more than 200kb) has been isolated against S. aureus; it has multi-domain RNA polymerase. Jumbo phages have multi-domain RNA polymerase. Many Gram-negative bacteria phages belong to this group; however, only a few Bacillus subtilis phages and two Staphylococcal phages have been identified (31). Regarding its physical robustness, the phage survival was assessed by placing it in different temperatures and pH. The most optimum condition for its activity is the room temperature and pH within a range of 7 - 8. Like most phages, ARW1 was stable at alkaline conditions but was inactivated in acidic conditions. However, the size of the plaque (zone of lysis) decreased in highly alkaline pH. The phage was stable at various temperatures (-20°C, 4°C, 25°C, and 37°C), but above 40°C, phage titre dropped sharply. The phage remained viable at -20°C for the entire period of this study (for three years). The phage was also active for a month at 4°C; Its titre did not diminish significantly.
We found that this phage has a fast adsorption rate, high burst size, and short latency period; which indicate its appropriateness as a candidate for biocontrol, as it can quickly adsorb and kill the host. Some examples of S. aureus phages with a short latency period are phage SPW (a Myoviridae phage) and phage SLPW (a Podoviridae phage) (32, 33). Regarding the host range, we tested it against environmental isolates of Gram-negative bacteria and S. aureus isolates, and it only produced clear plaques with isolate W1. This phage is specific and can be useful for MRSA detection in the environment. Our study was limited to environmental isolates. But its infectivity against various clinical and agricultural MRSA and S. aureus isolates can be tested; It may have a broad host range.
5.1. Conclusions
In general, isolating a phage against S. aureus is difficult. We got a phage against MRSA from the River Ganga. It is a Podoviridae phage; Podophages of S. aureus are rare and most desirable owing to their lytic lifecycle and lack of virulence as well as resistance genes. We present the first report of an S. aureus phage from the family Podoviridae that encodes its RNA polymerase.
References
-
1.
Rai A, Khairnar K. Overview of the risks of Staphylococcus aureus infections and their control by bacteriophages and bacteriophage-encoded products. Braz J Microbiol. 2021. [PubMed ID: 34251609]. https://doi.org/10.1007/s42770-021-00566-4.
-
2.
Rosenberg Goldstein RE, Micallef SA, Gibbs SG, He X, George A, Sapkota A, et al. Occupational exposure to Staphylococcus aureus and Enterococcus spp. among spray irrigation workers using reclaimed water. Int J Environ Res Public Health. 2014;11(4):4340-55. [PubMed ID: 24747541]. [PubMed Central ID: PMC4025025]. https://doi.org/10.3390/ijerph110404340.
-
3.
Boopathy R. Presence of methicillin resistant Staphylococcus aureus (MRSA) in sewage treatment plant. Bioresour Technol. 2017;240:144-8. [PubMed ID: 28262305]. https://doi.org/10.1016/j.biortech.2017.02.093.
-
4.
Rahimi F, Katouli M, Pourshafie MR. Characterization of methicillin-resistant Staphylococcus aureus strains in sewage treatment plants in Tehran, Iran. J Water Health. 2021;19(2):216-28. [PubMed ID: 33901019].
-
5.
Ju F, Li B, Ma L, Wang Y, Huang D, Zhang T. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters. Water Res. 2016;91:1-10. [PubMed ID: 26773390]. https://doi.org/10.1016/j.watres.2015.11.071.
-
6.
Mattila S, Ruotsalainen P, Jalasvuori M. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol. 2015;6:1271. [PubMed ID: 26617601]. [PubMed Central ID: PMC4643220]. https://doi.org/10.3389/fmicb.2015.01271.
-
7.
Latz S, Wahida A, Arif A, Hafner H, Hoss M, Ritter K, et al. Preliminary survey of local bacteriophages with lytic activity against multi-drug resistant bacteria. J Basic Microbiol. 2016;56(10):1117-23. [PubMed ID: 27194637]. https://doi.org/10.1002/jobm.201600108.
-
8.
Kaur S, Harjai K, Chhibber S. Methicillin-resistant Staphylococcus aureus phage plaque size enhancement using sublethal concentrations of antibiotics. Appl Environ Microbiol. 2012;78(23):8227-33. [PubMed ID: 23001655]. [PubMed Central ID: PMC3497353]. https://doi.org/10.1128/AEM.02371-12.
-
9.
Rakieten ML, Rakieten TL, Doff S. Absorption of Staphylococcus bacteriophages. J Bacteriol. 1936;32(5):505-18. [PubMed ID: 16559969]. [PubMed Central ID: PMC543816]. https://doi.org/10.1128/jb.32.5.505-518.1936.
-
10.
Deghorain M, Van Melderen L. The Staphylococci phages family: an overview. Viruses. 2012;4(12):3316-35. [PubMed ID: 23342361]. [PubMed Central ID: PMC3528268]. https://doi.org/10.3390/v4123316.
-
11.
Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA. 2005;102(14):5174-9. [PubMed ID: 15788529]. [PubMed Central ID: PMC556006]. https://doi.org/10.1073/pnas.0501140102.
-
12.
Oliveira H, Sampaio M, Melo LDR, Dias O, Pope WH, Hatfull GF, et al. Staphylococci phages display vast genomic diversity and evolutionary relationships. BMC Genomics. 2019;20(1):357. [PubMed ID: 31072320]. [PubMed Central ID: PMC6507118]. https://doi.org/10.1186/s12864-019-5647-8.
-
13.
Adams MH. Bacteriophages. Interscience Publishers; 1959.
-
14.
Jensen KC, Hair BB, Wienclaw TM, Murdock MH, Hatch JB, Trent AT, et al. Isolation and host range of bacteriophage with lytic activity against methicillin-resistant Staphylococcus aureus and potential use as a fomite decontaminant. PLoS One. 2015;10(7). e0131714. [PubMed ID: 26131892]. [PubMed Central ID: PMC4488860]. https://doi.org/10.1371/journal.pone.0131714.
-
15.
Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol. 2009;501:69-76. [PubMed ID: 19066811]. https://doi.org/10.1007/978-1-60327-164-6_7.
-
16.
Pickard DJ. Preparation of bacteriophage lysates and pure DNA: Bacteriophages. New York, USA: Humana press; 2009. p. 3-9.
-
17.
Ackermann HW. Basic phage electron microscopy: Bacteriophages. New York, USA: Humana Press; 2009. p. 113-26.
-
18.
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. [PubMed ID: 20211023]. [PubMed Central ID: PMC2848648]. https://doi.org/10.1186/1471-2105-11-119.
-
19.
McNair K, Bailey BA, Edwards RA. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics. 2012;28(5):614-8. [PubMed ID: 22238260]. [PubMed Central ID: PMC3289917]. https://doi.org/10.1093/bioinformatics/bts014.
-
20.
Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44(W1):W54-7. [PubMed ID: 27174935]. [PubMed Central ID: PMC4987944]. https://doi.org/10.1093/nar/gkw413.
-
21.
Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517-25.
-
22.
Kropinski AM. Measurement of the rate of attachment of bacteriophage to cells: Bacteriophages. New York, USA: Humana Press; 2009. p. 151-5.
-
23.
Ellis EL, Delbruck M. The growth of bacteriophage. J Gen Physiol. 1939;22(3):365-84. [PubMed ID: 19873108]. [PubMed Central ID: PMC2141994]. https://doi.org/10.1085/jgp.22.3.365.
-
24.
Hyman P, Abedon ST. Practical methods for determining phage growth parameters: In Bacteriophages. New York, USA: Humana Press; 2009.
-
25.
Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119-20. [PubMed ID: 18990721]. [PubMed Central ID: PMC2612626]. https://doi.org/10.1093/bioinformatics/btn578.
-
26.
Khairnar K. Ganges: special at its origin. J Biol Res (Thessalon). 2016;25(1):119-20. [PubMed ID: 27429942]. [PubMed Central ID: PMC4946093]. https://doi.org/10.1186/s40709-016-0055-6.
-
27.
Jofre J, Lucena F, Blanch A, Muniesa M. Coliphages as model organisms in the characterization and management of water resources. Water. 2016;8(5). https://doi.org/10.3390/w8050199.
-
28.
King AM, Lefkowitz E, Adams MJ, Carstens EB. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. 9. Elsevier; 2011.
-
29.
Culbertson EK, Bari SMN, Dandu VS, Kriznik JM, Scopel SE, Stanley SP, et al. Draft genome sequences of Staphylococcus podophages JBug18, Pike, Pontiff, and Pabna. Microbiol Resour Announc. 2019;8(8). [PubMed ID: 30834363]. [PubMed Central ID: PMC6386564]. https://doi.org/10.1128/MRA.00054-19.
-
30.
Catalao MJ, Gil F, Moniz-Pereira J, Sao-Jose C, Pimentel M. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol Rev. 2013;37(4):554-71. [PubMed ID: 23043507]. https://doi.org/10.1111/1574-6976.12006.
-
31.
Lee Y, Son B, Cha Y, Ryu S. Characterization and genomic analysis of pals2, a novel Staphylococcus Jumbo bacteriophage. Front Microbiol. 2021;12:622755. [PubMed ID: 33763042]. [PubMed Central ID: PMC7982418]. https://doi.org/10.3389/fmicb.2021.622755.
-
32.
Li L, Zhang Z. Isolation and characterization of a virulent bacteriophage SPW specific for Staphylococcus aureus isolated from bovine mastitis of lactating dairy cattle. Mol Biol Rep. 2014;41(9):5829-38. [PubMed ID: 24981924]. https://doi.org/10.1007/s11033-014-3457-2.
-
33.
Wang Z, Zheng P, Ji W, Fu Q, Wang H, Yan Y, et al. SLPW: A virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Front Microbiol. 2016;7. https://doi.org/10.3389/fmicb.2016.00934.