Abstract
Background:
Based on the WHO, multidrug-resistant Klebsiella pneumoniae is a priority pathogen that causes opportunistic infections and is widely spread in the environment. Phage therapy is considered a natural, safe, and very efficient alternative to treat difficult-to-treat infections.Objectives:
This study aimed to isolate highly virulent, lytic bacteriophages and evaluate their efficacy for lysing multidrug-resistant K. pneumoniae.Methods:
Municipal wastewater samples were collected and filtered using 0.22 µm syringe filters and cultivated with log-phase cultures of K. pneumoniae using enrichment media. After 48 h of incubation, the cultures were centrifuged, and the resultant supernatant was filtered (0.22 µm). The detection of the phage was done using the spot assay with K. pneumoniae as the host. One-step growth kinetics and bacterial reduction tests were conducted to assess the growth kinetics of the isolated phage. The stability of the isolated phage was characterized by subjecting it to various temperature and pH conditions. The chemical stability of the K. pneumoniae phage was determined by exposing it to various organic compounds. A panel of 20 bacterial strains was tested using the spot assay, as well as double agar overlying assay, to determine the host range of the isolated phage.Results:
Out of 40 wastewater samples tested, only one sample was tested positive for the K. pneumoniae phage (2.5%) that was lytic against the host strain. The K. pneumoniae phage had a latent period of 15 min and a burst size of 100 virions per infected cell. It was most stable at 37°C and pH range of 6.0 to 10.0. Chemically, the K. pneumoniae phage was resistant to 10% chloroform treatment. Transmission electron micrograph indicated that the K. pneumoniae phage belonged to the order Caudovirales, family Siphoviridae, morphotype B1.Conclusions:
Most of the characteristic features of the K. pneumoniae phage indicated the potential of this phage to be used in phage therapy. Hence, a comprehensive study is highly recommended to characterize the K. pneumoniae phage genome, detect its molecular interactions with the host cell, and determine its lytic activity in combination with other phages, which may lead to the efficient utilization of this phage in phage therapy against K. pneumoniae infections.Keywords
Klebsiella pneumoniae Bacteriophage Wastewater Multidrug-resistant
1. Background
Resistance to antimicrobial agents is a paradigm of evolution. Antimicrobial resistance develops as a consequence of misuse, abuse, and / or excessive use of antibiotics. The collective effect of different mechanisms of resistance leads to the development of multidrug-resistant bacterial pathogens, which may result in life-threatening infections that are extremely hard to treat. Nowadays, the rise of multi-drug resistant bacterial pathogens is of global public health concern, particularly in hospital settings (1). One of the most important bacteria is multidrug-resistant Klebsiella pneumoniae (2). Multidrug-resistant (MDR) K. pneumoniae is considered by the WHO as a priority pathogen, which causes opportunistic and nosocomial infections in hospital settings and has a broad environmental distribution. Klebsiella pneumoniae is the principal cause of hospital-acquired infections such as bloodstream infections, pneumonia, newborns’ infection, and infections associated with immunocompromised patients (3). Besides, community-acquired pyogenic liver abscess caused by MDR K. pneumoniae convoluted with endophthalmitis and metastatic meningitis has emerged worldwide, particularly in Asia (4). Many K. pneumoniae strains have been recognized to be unresponsive to the last resort of antimicrobial agents like carbapenem, fluoroquinolones, cephalosporin, and colistin (2).
Phage therapy is considered a natural, safe, and very efficient alternative to counteract the increasing problems associated with multidrug-resistant bacterial infections (5). Individual lytic phages, as well as cocktails of phages targeting different pathogenic bacteria, including K. pneumoniae, Enterococcus spp., Pseudomonas aeruginosa, Escherichia coli, staphylococci, and other pathogenic bacterial strains, have been successful at controlling the bacterial density in in vivo and in vitro experiments (6-8). Phage therapy could provide substantial protection against respiratory and other infections caused by K. pneumoniae, such as bacteremia and liver abscesses in the mice model (9). Phage therapy has also been used to cure K. pneumoniae-infected burn wounds in mice (10). Intranasal delivered lytic phage reduced the load of K. pneumoniae in the lung of mice (11, 12). Recently, a significant reduction of the bacterial load has been reported by using a lytic Myoviridae phage, vB_KpnM- Teh, after an intranasal injection of K. pneumoniae to BALB/c mice (13). Several other studies have identified different types of lytic bacteriophages against K. pneumoniae and investigated their efficacy in in vitro and in vivo studies (14, 15).
Despite the enormous benefits of phage therapy, the evolution of phage resistance poses an inevitable threat to the efficacy of phage therapy. Currently, it has been reported that phage-resistant K. pneumoniae mutants became unresponsive to specific phage treatments (16). Hence, isolating a new potently lytic bacteriophage is very crucial in combating MDR and phage-resistant bacterial infections simultaneously.
2. Objectives
This study aimed to isolate highly virulent, lytic bacteriophages and evaluate their efficacy in lysing MDR K. pneumoniae.
3. Methods
3.1. Bacterial Isolates and Culture Conditions
The clinical strains of K. pneumoniae were obtained from King Abdulaziz University Hospital. The identification of the bacterial strain was conducted by cultural characteristics (shape and morphology of its colony, odor, and pigment production), Vitek 2 and API-20E (BioMerieux Industry, Hazelwood, MO, USA) bacterial identification tools (BioMarieux, Inc. Durham, NC). Lastly, the colonies were confirmed by 16s rRNA sequencing using specific (universal) primers. Purified colonies were picked up from the nutrient agar plate and grown on nutrient broth (Oxoid, ® Hampshire, England). After overnight incubation, the bacterial suspension was preserved in 50% (v/v) glycerol and kept at -20°C for further use.
3.2. Antibiotic Susceptibility Test
Drug sensitivity tests were conducted using the Kirby-Bauer disc diffusion method on Mueller-Hinton agar media according to the CLSI (2014) guidelines (17). Hi-media antibiotic discs tested were ciprofloxacin (CIP; 5 μg), gentamicin (CN; 10 μg), piperacillin (PRL; 100 μg), amikacin (AK; 30 μg), tobramycin (TOB; 10 μg), neomycin (NEO; 15 µg), streptomycin (STR; 5 µg), cefotaxime (CTX; 30 μg), cefuroxime (CXM; 30 μg), ciprofloxacin (CIP; 5 μg), imipenem (IPM; 10 μg), ceftriaxone (CRO; 30 μg), ticarcillin (TCR; 75 μg), ceftazidime (CAZ; 30 μg), levofloxacin (LEV; 5 μg), colistin (10 μg), and meropenem (MEM; 10 μg). In this test, the K. pneumonia ATCC 700603 strain was used as a positive control. Those K. pneumoniae strains that were resistant to all antimicrobial agents were considered Pan-drug-resistant (PDR). However, if they were resistant to at least one drug in three or more antimicrobial categories, they were considered MDR. Likewise, those isolates that were non-susceptible to at least one agent in two or fewer categories were characterized as Extensive drug-resistant (XDR) isolates (18).
3.3. Wastewater Sample Collection and Transportation
The study was conducted between November 2019 and March 2020. During the study period, a total of 40 different raw wastewater samples (each 20 mL) were collected from four different sites located in Jeddah, Saudi Arabia (Jeddah wastewater treatment plant, Al-Nakalile, Al-Nawras, and Al- Arbeen districts). The sample collection sites were selected based on the level of the contamination of water samples and the proximity of the site to our laboratory. All samples were collected in clean screw cap bottles and transported to the King Fahd Medical Research Center, Jeddah, Saudi Arabia (KFMRC), in an icebox. The collected samples were kept in a refrigerator until processed within 24 h of collection (19). All of the samples were assessed individually for the existence of lytic bacteriophages using PDR K. pneumoniae as a host organism.
3.4. Host Bacterial Strains
Pan drug-resistant K. pneumoniae 7 was used as a host organism for the isolation of bacteriophages from municipal wastewater samples.
3.5. Enrichment and Isolation of Bacteriophages
The isolation and enrichment of the phages were conducted based on (20) with minor modifications. Briefly, 20 mL of wastewater sample was centrifuged at 9,500 × g (700 rpm) for 12 min to remove any debris and suspended solids. The supernatant (20 mL) was transferred to a sterile 250 mL Erlenmeyer flask containing 20 mL of 24 h host grown culture and an equal volume of double-strength nutrient broth supplemented with 2 mM CaCl2. The mixture was incubated for 24 - 48 h with slight shaking at 100 rpm, 37°C. Thereafter, the suspension was spun down at 9,500 × g for 12 min at 4°C, and the supernatant was filtered with 0.22 µm syringe filters (Fischer Scientific, Ottawa, ON, Canada) to remove bacterial debris. Finally, the filtrates were stored at 4°C.
The spot assay was used to determine the lytic activity of phages against the host strains (21). Briefly, 100 µL of an exponential phase bacterial culture grown in fresh nutrient broth was mixed with 5 mL of molten soft agar (0.7% w/v agar) held at 55ºC in a warm water bath and then poured quickly onto sterile solid nutrient agar plates to form a lawn of the host bacteria. The bacteriolytic activity of the phage lysate was tested by spotting 10 µL of the crude extract (1 × 107 PFU/mL) on the top of soft agar and incubated for 20 min to allow viral adsorption. Plates were labeled and incubated at 37ºC for 24 h. Thereafter, each plate was observed for the presence of the defined lytic zones at the spots where the lysate was pipetted. The phages that produced a clear plaque were considered the virulent phages and were selected for further analysis.
3.6. Purification of Phages
The purification of phages was conducted using the soft agar overlay method as described by (22). Shortly, the plates that were positive for plaques (dispersed plaques) were selected, and one plaque was picked up from the top layer by touching them using a 1-mL pipette tip and placed separately in 2-mL Eppendorf tubes containing 500 µL Phosphate-buffered Saline (PBS) solution (pH 7.4). The suspension was incubated overnight at 4ºC to allow the phages to diffuse out from the soft agar into the suspension. Three successive experiments were conducted until getting single plaques with similar morphologies. Lastly, a 10-fold serial dilution was prepared, and the double-layered technique was used to determine the phages’ titers. The purified phage filtrate was stored at 4ºC for further use.
3.7. Phage Stability Tests
3.7.1. Temperature
The thermal stability of the K. pneumoniae phage was carried out according to the method described by (23). Shortly, 100 µL phage lysates (108 PFU/mL) were suspended in different sterile Eppendorf tubes containing an equal volume of PBS (pH 7.4) and incubated in water baths set at different temperatures ranging between 37ºC and 90ºC for 60 min. Then, the phage titer was determined using the double agar layer assay. The percentage stability was calculated by the ratio of surviving phage titers after treatment to the phage titer before treatment, multiplied by 100. The thermal stability curve (the mean of triplicate experiments versus time) was plotted using GraphPad Prism.
3.7.2. pH
The stability of the K. pneumoniae phage to different thermal treatments was conducted based on (24) with a very minor modification. Concisely, 1 M HCl and NaOH were added dropwise into the fresh nutrient broth and set at different pH values. Nine milliliters of pH-adjusted media were mixed with one-milliliter phage suspension and incubated for 2 h at 37ºC. The phage titer of each suspension was assessed by the double-layer agar method. The percentage stability was calculated by the ratio of surviving phage titers after treatment to the phage titer before treatment, multiplied by 100. The pH stability curve (the mean of triplicate experiments versus time) was plotted using GraphPad Prism.
3.7.3. Organic Solvents
The stability of the K. pneumoniae phage was assessed in the presence of organic solvents as indicated by (23). Briefly, 1,000 µL (6 × 107 PFU/mL) of phage suspension was mixed with an equal volume of organic solvents (10% chloroform, 70%, and 99% ethanol, and isopropanol) separately in sterile tubes and incubated at 37ºC for 2 - 8 h with gentle shaking. Thereafter, the mixtures were centrifuged at 9,500 × g for 12 min, and the phage titer in the aqueous phase was determined by the double-layer agar method. An equal volume of the phage lysate suspended in suspension mixture buffer was used as a control.
3.8. One-round Growth Pattern
A single-round growth pattern was done to assess the burst size and the latent period as stated by (21), with minor modifications. Briefly, 6 mL of the host cell suspension in the mid-exponential phase (OD600 = 0.4) was spun down at 9,500 × g for 5 min. The pelleted cell mass was re-suspended in 300 µL of fresh nutrient broth medium, and then 600 µL of purified lytic phage suspension was added to get a 0.01 MOI. Five minutes later, the mixture was spun down at 9,500 × g for 12 min to remove any un-adsorbed phages. Then, the pellet was re-suspended in 6,000 µL of fresh nutrient broth medium and incubated with shaking (100 rpm min−1) at 37ºC for 24 h. The aliquots were collected at 10 min intervals over a 40 min period. These aliquots were diluted with the phage buffer, and the phage titers were determined using the soft agar overlying method. These experiments were carried out at least in triplicate.
3.9. Bacterial Reduction Assay
The in vitro lytic activity of the purified phage was assessed as formerly stated by (25) with minor modifications. Shortly, 100 µL of the concentrated phage lysate (1 × 109 PFU/mL) was mixed with an equal volume of broth culture at MOI values of 100, 10, 1, and 0.1 in a 96 well-microliter plate and incubated at 37ºC. Then, the bacterial reduction rate was assessed by measuring the OD at 600 nm at one-hour intervals for 30 h using a multimode microplate reader (Tecan Spark 10M, USA). One well-containing log phage bacterial culture and PBS (equal proportions) served as a negative control. The results are displayed as mean ± Standard Deviation (SD) from three individual experiments, and the growth curve (OD600 vs. time) was plotted using GraphPad Prism.
3.10. Phage Host Range
The host range of the lytic phage isolated in this study was determined by the spot test described by (23) in a panel of 20 bacterial strains belonging to different genera (Table 1). Shortly, 5 µL of the purified lytic phage lysate (108 PFU/mL) was spotted onto the lawn of the selected host bacterium, which was produced by mixing 1 mL of overnight culture with 0.7% molten soft agar. It was ultimately poured over the surface of a modified nutrient agar plate. After overnight incubation, the lytic ability of the phage lysate was determined by the presence of clear plaques produced. According to the presence and absence of plaques, the results were classified into two categories: (−) no plaques and (+) positive-clear plaques.
Host Range of Klebsiella pneumoniae Phage Against 20 Bacterial Isolates
| Bacterial Strains | Source | Spot Test Result a | Confirmation with Plaque Assay a |
|---|---|---|---|
| Escherichia coli | ATCC25922 | - | - |
| Streptococcus progenies | ATCC19615 | - | - |
| S. bovis | ATCC49147 | - | - |
| S. pneumoniae | ATCC49619 | - | - |
| Hemophilus influenzae | ATCC49766 | - | - |
| Enterobacter cloacae | ATCC23355 | - | - |
| E. faecalis | ATCC29212 | - | - |
| Shigella sonnei | ATCC25931 | - | - |
| Klebsiella oxytoca | ATCC49131 | + | + |
| Staphylococcus saprophyticus | ATCC15305 | - | - |
| Moraxella catarrhalis | ATCC16605 | - | - |
| K. pneumonia clinical isolate | KFMRC | + | + |
| S. aureus clinical isolate | KFMRC | - | - |
| S. sonnei clinical isolate | KFMRC | - | - |
| Salmonella typhi clinical isolate | KFMRC | - | - |
| E. faecalis clinical isolate | KFMRC | - | - |
| S. agalactia | ATCC12386 | - | - |
| S. epidermidis | ATCC12281 | - | - |
| Proteus vulgaris | ATCC49132 | - | - |
| S. aureus | ATCC25923 | - | - |
| Pseudomonas aeruginosa | ATCC27853 | - | - |
| K. pneumonia | ATCC700603 | + | + |
| H. parainfluenza | ATCC79011 | - | - |
| MRSA | ATCC43330 | - | - |
3.11. Transmission Electron Microscopy
The K. pneumoniae phage was prepared for transmission electron microscopic (TEM) analysis by adhering the virion onto the carbon film, stained with the negative staining technique with uranyl acetate (2% (w/v), pH 5.0) (26). The concentrated phage lysate (1 × 108) was observed using a TEM 910 microscope (Carl Zeiss, Oberkochen, Germany) at 80 kV acceleration voltage at King Abdulla Science and Technology University, Jeddah, Saudi Arabia. Images were captured at optimal magnifications. The phage morphology was determined using the benchmarks defined by the International Committee of Taxonomy of Viruses (ICTV). The diameter of the head and tail of the captured phage image was measured using Image J software. Lastly, we classified the phage based on the criteria proposed by (27).
3.12. Statistical Analysis
The data were recorded as mean ± SD. GraphPad Prism version 6 (GraphPad Software Inc. USA) was used to analyze the data. The Kolmogorov-Smirnov test was used to check the normal distribution of the data. A Student’s t-test or one-way analysis of variance with post hoc Tukey’s multiple comparison test was used to determine differences between two or multiple groups, respectively. Besides, P < 0.05 (∗) was considered statistically significant.
4. Results
4.1. Characteristics of Klebsiella pneumoniae
A total of seven clinical strains of K. pneumoniae (7/30) were obtained from King Abdulaziz University Hospital, Jeddah, Saudi Arabia. The isolated strains were identified by colony characteristics, Vitek 2, and PAI-20E systems. The bacterial isolates showed two distinct colony characteristics. The first six isolates showed dry, rough, and transparent colonies, whereas the last one (K. pneumoniae 7) showed mucoid, moist, and sticky colonies in in vitro cultivation (Table 2). The seven-digit profile number (2212000) obtained from the API-20E test indicated that the organism was 97% identical to K. pneumoniae. In addition, the partial sequence of the 16S rRNA gene revealed that K. pneumoniae 7 was 100% identical to K. pneumoniae (Accession no. AP025038.1).
Morphological and Drug Resistance Profiles of Klebsiella pneumoniae Isolates
| Isolates | Colony Characteristics | Patterns of Drug Resistance | Level of Resistance | Remarks |
|---|---|---|---|---|
| Klebsiella pneumoniae 1 | Dry, rough, and transparent | CN, CAZ, CAP | MDR | - |
| K. pneumoniae 2 | Dry, rough, and transparent | CN, CAZ, CAP | MDR | - |
| K. pneumoniae 3 | Dry, rough, and transparent | CN, CAZ, CAP | MDR | - |
| K. pneumoniae 4 | Dry, rough, and transparent | CN, CAZ | XDR | - |
| K. pneumoniae 5 | Dry, rough, and transparent | CN, CAZ | XDR | - |
| K. pneumoniae 6 | Dry, rough, and transparent | CN, CAZ | XDR | - |
| K. pneumoniae 7 | Mucoid, moist, and sticky | All tested antibiotics including Carbapenem and Colistin | PDR | Selected for phage isolation |
4.2. Antibiotic Sensitivity Test
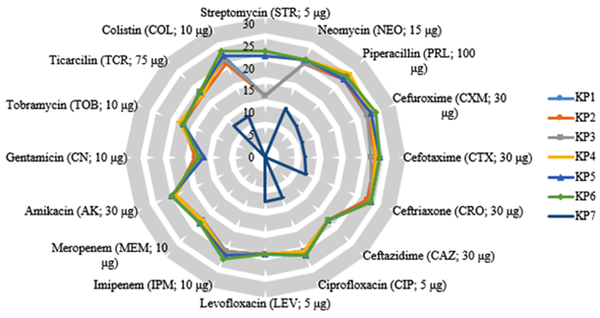
The antimicrobial sensitivity results indicated that three isolates (K. pneumoniae 1, 2, and 3) were resistant to three antibiotics (Table 2) whereas three isolates (K. pneumoniae 4, 5, and 6) were resistant to two antibiotics. However, one isolate (K. pneumoniae 7) was resistant to almost all of the tested antibiotics, which was later used as the host organism for the isolated phage from wastewater (Figure 1).
Radar plot of antibiotic susceptibility profiles of Klebsiella pneumoniae isolates. The inhibition zones were recorded in mm.
4.3. Isolation of Phage, Plaque, and Virion Morphology
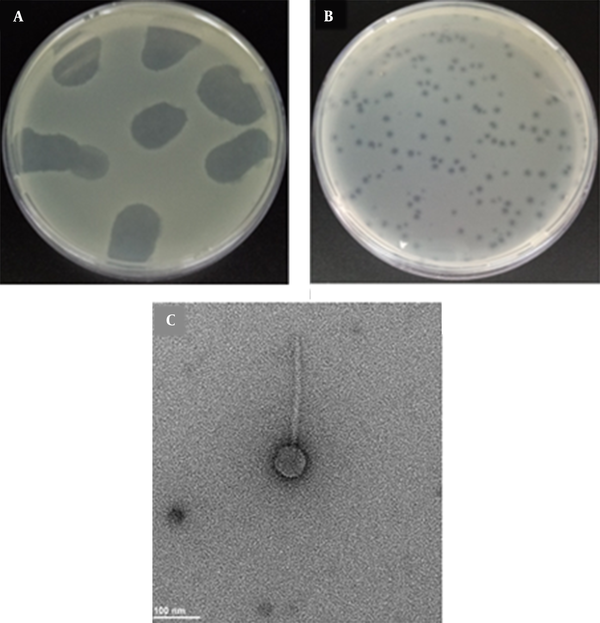
A total of 40 municipal wastewater samples were screened separately for the presence of bacteriophages using MDR K. pneumoniae 7 as the host. Out of 40 samples tested, only one was tested positive for the phage (2.5%) that was lytic against the host strain. The morphology of K. pneumoniae phage plaques was determined by the double agar overlying test. Small, clear, round, and non-halo plaques with an average diameter of ≤ 1 ± 0.2 mm were recorded (Figure 2B). The TEM images indicated that the K. pneumoniae phage had a long, non-contractile tail (145 ± 1.2 nm length) with transverse striation, icosahedral head, and non-prolate capsid (57 ± 1.2 nm diameter). The diameter of the tail was found to be 12 nm (Figure 2C). No tail fibers were observed in the TEM images. According to ICTV, the observed morphology indicated that the phage was a member of the Siphoviridae family, B1 morphotype.
Bacteriophage isolated against multidrug-resistant Klebsiella pneumoniae. A, Spot assay on a lawn of MDR K. pneumoniae; B, Plaques of K. pneumoniae phage formed on a lawn of MDR K. pneumoniae using double-layer agar technique; C, Transmission electron micrograph of K. pneumoniae phage. The bar represents 100 nm.
4.4. One-step Growth
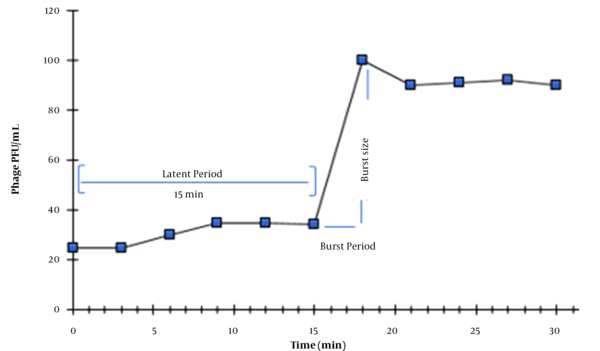
The single-round growth cycle analysis was performed to assess the latent period and burst size of the K. pneumoniae phage as presented in Figure 3. A growth curve with three phases (latent phase, log phase, and stationary phase) was obtained. Accordingly, the burst size of the K. pneumoniae phage was found to be 100 phages per infected cell with a latent period of 15 min.
One-step growth curve of Klebsiella pneumoniae phage. Values are the means of triplicate assays. Note that error bars are not visible due to their small sizes.
4.5. Bacterial Challenge Test
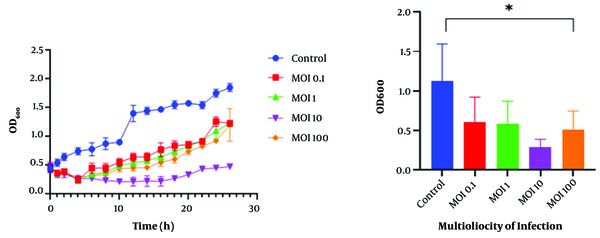
The bacterial challenge assay showed that the bacterial cells’ growth was inhibited in the presence of the K. pneumoniae phage at different MOIs. The growth of the host bacteria infected with the K. pneumoniae phage was reduced for 4 h at MOI 0.1, 1, and 100, and was efficiently inhibited for 16 h at MOI 10 (optimal multiplicity of infection). In contrast, normal growth was observed at MOI 0 (control) and the optical density (OD600) increased continuously during the incubation period (Figure 4).
Bacterial challenge test of Klebsiella pneumoniae phage. Effects of MOI 0.1, MOI 1, MOI 10, and MOI 100 of K. pneumoniae phage on the growth of log-phase culture of K. pneumoniae compared with a control culture (without phage) (* P < 0.05). Values are mean ± SD taken from triplicate assays.
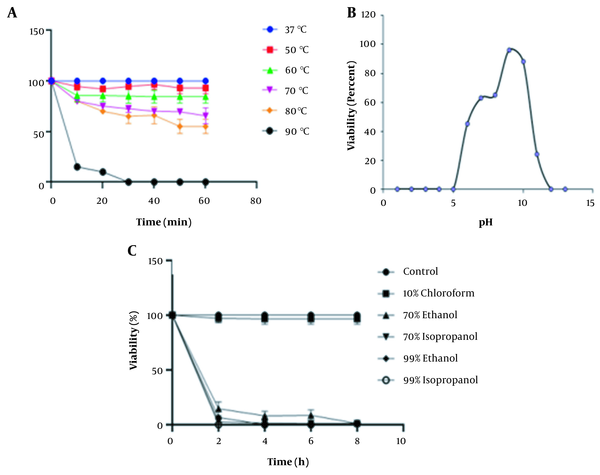
4.6. Stability of K. pneumoniae Phage to Thermal, pH, and Organic Solvent Treatments
The thermal stability test showed that the K. pneumoniae phage was relatively stable at temperatures ranging from 37 to 60°C for 60 min. However, the K. pneumoniae phage survival rate steadily declined at 90°C where it reached zero after 30 min of incubation (P < 0.05) (Figure 5A). In addition, we found that greater stability of the K. pneumoniae phage was recorded between pH 6 and pH 10 for at least 2 h. However, the K. pneumoniae phage was fully inactivated at pH ≤ 5 and ≥ 12 (P < 0.05) (Figure 5B). Chemically, the K. pneumoniae phage was stable when treated with 10% chloroform. However, we observed that the survival rate of the K. pneumoniae phage was adversely affected in the presence of 70% and 99% ethanol and isopropanol treatment (P < 0.05) (Figure 5C).
A, Thermal stability of Klebsiella pneumoniae phage at 10 min intervals for 60 min; B, pH stability test; C, The percentage viability of K. pneumoniae phage against various organic solvents. Values are mean ± SD taken from triplicate assays. Note that error bars are not visible due to their small sizes.
4.7. Host Range
The host range of the lytic phage isolated in this study was determined by the spot assay on a panel of 20 strains belonging to different genera (Table 1). The spot test results indicated that the K. pneumoniae phage had no lytic activity against the non-targeted bacterial strains tested. The lytic activity of the K. pneumoniae phage was specific to clinical isolates of K. pneumoniae, K. oxytoca (ATCC 49131), and K. pneumoniae (ATCC 700603). During the cultivation process of K. pneumoniae coupled with its phage, a re-growth of bacteria (phage-resistant mutants) was noticed in the lytic zone in a few culture plates mostly 48 h post-incubation. The bacterial colonies colonizing the lytic zone were sub-cultured in Cysteine-lactose-electrolyte-deficient (CLED) agar and showed distinct characteristics of K. pneumoniae. The API-20E test indicated that the organism was K. pneumoniae.
5. Discussion
Currently, the spread of MDR K. pneumoniae poses a serious threat to public health globally (28). In this study, we identified different K. pneumoniae isolates from samples obtained from King Abdulaziz University Hospital. The antimicrobial sensitivity test results indicated that one of the isolates (K. pneumoniae 7) was resistant to a wide range of antibiotics including carbapenem and colistin. This finding is in harmony with the former findings, which indicated that K. pneumoniae was the most frequently isolated strain from patients treated in hospitals (2). Moreover, several studies indicated that hospitals and/or clinics are the potential sites for emerging MDR bacterial strains (29, 30). Bacteriophages are promising alternative therapeutic agents against multidrug-resistant bacterial pathogens (31). In this study, a lytic bacteriophage against MDR K. pneumoniae was isolated from municipal wastewater samples. Similarly, in a study by Kumari et al., five bacteriophages (K. pneumoniae 5, 12, 13, 17, and 22), lytic against K. pneumoniae strain B5055, were isolated from sewage samples (32). Recently, a potentially lytic Myoviridae phage, vB_KpnM-Teh.1, was also isolated from wastewater samples, which exhibited its efficacy against a clinical strain of K. pneumoniae (13).
The isolated K. pneumoniae phage had a unique three-phase growth pattern: latent phase, exponential phase, and stationary phase. The growth profile of this phage showed a moderate latent period (15 min) and burst size (100 virions/infected cell). A former study on a Klebsiella species-infecting phage belonging to the Podoviridae family showed nearly similar growth patterns with a latent period of 15 min and burst size of ≈ 50 – 60 virions / infected cell (33). A similar latent period (15 min) was reported by others for the K. pneumoniae phage, KP34 (φKMV-like bacteriophage), isolated from sewage samples. The burst size of this phage was ∼40 – 50 virions/infected cell (34). However, this was not the case of the Myoviridae K. pneumoniae phage, vB_KpnM_KP27, isolated from the communal wastewater treatment plant, which revealed a lower burst size of 10 – 15 PFU per infected cell and a longer latent period (25 min) (35). Dalmasso et al. reported that the burst size of phages mainly depends on the number of susceptible host cells available (36). Santos et al. confirmed that the lytic activity of phages is determined by the latent period, as well as the burst size of the phage (37). The shortest latent period accompanied by the largest burst size led to a quick phage replication cycle, which could result in high progeny virions released outside from the infected host cells (38).
The stability of the K. pneumoniae phage was studied at different pH values to obtain very crucial data on its suitability for phage therapy. The results obtained related to the effect of pH on K. pneumoniae phage stability agreed with those of Anand et al. on phage BPA43, which showed moderate stability between pH 4 and 10 (11). The wide or moderate pH range tolerance of phages is advantageous when used in oral phage therapy after passing the clinical trials (39, 40). The results on thermal stability were comparable with those of Wintachai et al., who reported that the Siphoviridae K. pneumoniae phage, K. pneumoniae 1801, showed stability over a wide range of temperature from 80 to 60°C (41). Contrary, the K. pneumoniae phage Bp5, was found to be active at temperatures not exceeding 50°C (42). The variations in thermal phage stability could be due to differences in strains, structures of the phage, or the sites of phage isolation (43). However, most studies have indicated that phage viability is affected by high temperatures (44, 45).
The results on the effects of organic compounds on K. pneumoniae phage stability are in parallel with those of Pallavi et al., indicating that phage vB-AhyM-AP1 showed stability after treatment with chloroform for 10 min incubation (46). Concerning the bacterial killing assay, a high level of K. pneumoniae reduction was noted at an MOI of 10, which is consistent with what was mentioned elsewhere (47). The K. pneumoniae phage showed a narrow host range, lysing only 55.5% (3 out of 20) of bacterial isolates. Similar to our results, Soleimani Sasani et al. reported a Siphoviridae phage, designated as vB_KpnS-Teh.1, which showed a narrow range of lytic activity against K. pneumoniae, and lysed only 15.7% (8 out of 51) of extended-spectrum beta-lactamase-positive isolates (48). In other studies, the members of Siphoviridae were reported to exhibit similar activity against K. pneumoniae, lysing 7 - 15% of MDR isolates (35, 49-51). These phages multiply more efficiently and show faster elimination of their bacterial hosts (35). The narrow host range can be advantageous in phage therapy since it lowers the possibility of affecting other members of the normal flora. Contrary, the narrow activity spectrum limits the application of the isolated phages to very specific bacterial infections. However, this problem can be solved by using different phages together as a cocktail, which broadens the range of bacteriolytic activity against the targeted and non-targeted bacterial pathogens (52).
In our study, we detected phage-resistant mutant bacterial colonies after overnight incubation of the K. pneumoniae phage with the host bacterial strain. This finding agrees with the findings of Hesse et al., who reported that a total of 57 phage-resistant K. pneumoniae mutants evolved after prolonged co-culture in vitro (16). The re-growth of bacteria after phage treatment is mostly associated with the emergence of phage-resistant mutants, which complicates the clinical trials of phage therapy (53). This phenomenon has been attributed to phenotypic and genetic changes, which allow bacterial cells to maintain their viability in the presence of phages (54, 55). Phage resistance is rarely encountered in in vivo conditions as compared to in vitro environments, which could be due to the immune system that scavenges the mutants (56). Several research findings have revealed that the development of phage resistance can be mitigated by using phage cocktails and/or phage antibiotic combinations. However, the reduction rate or the effectiveness of these approaches depends on the efficiency of the phage and the potency of the antibiotic selected (57, 58).
5.1. Conclusions
In conclusion, the lytic virulent phage, which was isolated against MDR K. pneumoniae, had a narrow host range spectrum, which makes it more advantageous with fewer adverse effects on the normal flora. The K. pneumoniae phage showed a good (moderate) survival rate in pH and thermal treatments. In addition, the latent period and the burst size of the K. pneumoniae phage indicated the possibility of its potential use in phage therapy. Based on that, it would be suggested that the in vivo activity of the K. pneumoniae phage be elucidated to better understand the interactions of the phage with the host organism and the underlying mechanisms by which it evades the host defense system. The K. pneumoniae phage should also be characterized at the molecular level, which will provide a more comprehensive insight into the biology of the phage. Besides, phage-resistant K. pneumoniae mutants have been encountered during our study. This phenomenon needs to be elucidated further. Future work should focus on the use of the isolated phage in combination with other potential lytic phages as a cocktail and / or with conventional antibiotics.
Acknowledgements
References
-
1.
Fiaz M, Ahmed I, Riaz R, Nawaz U, Arshad M. Prevalence of antibiotic-resistant bacterial strains in wastewater streams: molecular characterization and relative abundance. Folia Microbiol (Praha). 2021. [PubMed ID: 34339002]. https://doi.org/10.1007/s12223-021-00902-z.
-
2.
Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1-9. [PubMed ID: 31918737]. [PubMed Central ID: PMC7050612]. https://doi.org/10.1186/s12941-019-0343-8.
-
3.
Le T, Wang L, Zeng C, Fu L, Liu Z, Hu J. Clinical and microbiological characteristics of nosocomial, healthcare-associated, and community-acquired Klebsiella pneumoniae infections in Guangzhou, China. Antimicrob Resist Infect Control. 2021;10(1):1-11. [PubMed ID: 33632338]. [PubMed Central ID: PMC7908793]. https://doi.org/10.1186/s13756-021-00910-1.
-
4.
Shi YF, Wang YK, Wang YH, Liu H, Shi XH, Li XJ, et al. Metastatic infection caused by hypervirulent Klebsiella pneumonia and co-infection with Cryptococcus meningitis: A case report. World J Clin Cases. 2019;7(22):3812-20. [PubMed ID: 31799309]. [PubMed Central ID: PMC6887595]. https://doi.org/10.12998/wjcc.v7.i22.3812.
-
5.
Taati Moghadam M, Khoshbayan A, Chegini Z, Farahani I, Shariati A. Bacteriophages, a new therapeutic solution for inhibiting multidrug-resistant bacteria causing wound infection: lesson from animal models and clinical trials. Drug Des Devel Ther. 2020;14:1867-83. [PubMed ID: 32523333]. [PubMed Central ID: PMC7237115]. https://doi.org/10.2147/DDDT.S251171.
-
6.
Al-Ishaq RK, Skariah S, Busselberg D. Bacteriophage treatment: critical evaluation of its application on world health organization priority pathogens. Viruses. 2020;13(1):51. [PubMed ID: 33396965]. [PubMed Central ID: PMC7823271]. https://doi.org/10.3390/v13010051.
-
7.
Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R. Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter species. Front Microbiol. 2019;10:574. [PubMed ID: 30949158]. [PubMed Central ID: PMC6437105]. https://doi.org/10.3389/fmicb.2019.00574.
-
8.
Broncano-Lavado A, Santamaria-Corral G, Esteban J, Garcia-Quintanilla M. Advances in bacteriophage therapy against relevant multi drug-resistant pathogens. Antibiotics (Basel). 2021;10(6):672. [PubMed ID: 34199889]. [PubMed Central ID: PMC8226639]. https://doi.org/10.3390/antibiotics10060672.
-
9.
Hung CH, Kuo CF, Wang CH, Wu CM, Tsao N. Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob Agents Chemother. 2011;55(4):1358-65. [PubMed ID: 21245450]. [PubMed Central ID: PMC3067190]. https://doi.org/10.1128/AAC.01123-10.
-
10.
Hesse S, Malachowa N, Porter AR, Freedman B, Kobayashi SD, Gardner DJ, et al. Bacteriophage treatment rescues mice infected with multidrug-resistant Klebsiella pneumoniae ST258. mBio. 2021;12(1). [PubMed ID: 33622728]. [PubMed Central ID: PMC8545083]. https://doi.org/10.1128/mBio.00034-21.
-
11.
Anand T, Virmani N, Kumar S, Mohanty AK, Pavulraj S, Bera BC, et al. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J Glob Antimicrob Resist. 2020;21:34-41. [PubMed ID: 31604128]. https://doi.org/10.1016/j.jgar.2019.09.018.
-
12.
Cao F, Wang X, Wang L, Li Z, Che J, Wang L, et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed Res Int. 2015:752930. [PubMed ID: 25879036]. [PubMed Central ID: PMC4387947]. https://doi.org/10.1155/2015/752930.
-
13.
Soleimani Sasani M, Eftekhar F. Potential of a bacteriophage isolated from wastewater in treatment of lobar pneumonia infection induced by Klebsiella pneumoniae in mice. Curr Microbiol. 2020;77(10):2650-5. [PubMed ID: 32451685]. https://doi.org/10.1007/s00284-020-02041-z.
-
14.
Hoyles L, Murphy J, Neve H, Heller KJ, Turton JF, Mahony J, et al. Klebsiella pneumoniae subsp. pneumoniae-bacteriophage combination from the caecal effluent of a healthy woman. PeerJ. 2015;3. e1061. [PubMed ID: 26246963]. [PubMed Central ID: PMC4525690]. https://doi.org/10.7717/peerj.1061.
-
15.
Herridge WP, Shibu P, O'Shea J, Brook TC, Hoyles L. Bacteriophages of Klebsiella spp., their diversity and potential therapeutic uses. J Med Microbiol. 2020;69(2):176. https://doi.org/10.7287/peerj.preprints.27890v1.
-
16.
Hesse S, Rajaure M, Wall E, Johnson J, Bliskovsky V, Gottesman S, et al. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. mBio. 2020;11(1). [PubMed ID: 31992617]. [PubMed Central ID: PMC6989104]. https://doi.org/10.1128/mBio.02530-19.
-
17.
Wayne PA. Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. 2010.
-
18.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-81. [PubMed ID: 21793988]. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
-
19.
Olson ME, Horswill AR. Bacteriophage Transduction in Staphylococcus epidermidis. 1106. Clifton, New Jersey: Methods Mol Biol; 2014. p. 167-72.
-
20.
Van Twest R, Kropinski AM. Bacteriophage enrichment from water and soil. Methods Mol Biol. 2009;501:15-21. [PubMed ID: 19066806]. https://doi.org/10.1007/978-1-60327-164-6_2.
-
21.
Pereira C, Silva YJ, Santos AL, Cunha A, Gomes NC, Almeida A. Bacteriophages with potential for inactivation of fish pathogenic bacteria: survival, host specificity and effect on bacterial community structure. Mar Drugs. 2011;9(11):2236-55. [PubMed ID: 22163184]. [PubMed Central ID: PMC3229233]. https://doi.org/10.3390/md9112236.
-
22.
Gencay YE, Birk T, Sorensen MC, Brondsted L. Methods for Isolation, Purification, and Propagation of Bacteriophages of Campylobacter jejuni. 1512. Clifton, New Jersey: Methods Mol Biol; 2017. p. 19-28.
-
23.
Jurczak-Kurek A, Gasior T, Nejman-Falenczyk B, Bloch S, Dydecka A, Topka G, et al. Biodiversity of bacteriophages: morphological and biological properties of a large group of phages isolated from urban sewage. Sci Rep. 2016;6:34338. [PubMed ID: 27698408]. [PubMed Central ID: PMC5048108]. https://doi.org/10.1038/srep34338.
-
24.
Verma V, Harjai K, Chhibber S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: A potential therapeutic agent. Curr Microbiol. 2009;59(3):274-81. [PubMed ID: 19484297]. https://doi.org/10.1007/s00284-009-9430-y.
-
25.
Wang Z, Zheng P, Ji W, Fu Q, Wang H, Yan Y, et al. SLPW: A virulent bacteriophage targeting methicillin-resistant Staphylococcus aureus in vitro and in vivo. Front Microbiol. 2016;7. https://doi.org/10.3389/fmicb.2016.00934.
-
26.
Valentine RC, Thang MN, Grunberg-Manago M. Electron microscopy of Escherichia coli polynucleotide phosphorylase molecules and polyribonucleotide formation. J Mol Biol. 1969;39(2):389-91. https://doi.org/10.1016/0022-2836(69)90326-x.
-
27.
Ackermann HW. 5500 Phages examined in the electron microscope. Arch Virol. 2007;152(2):227-43. [PubMed ID: 17051420]. https://doi.org/10.1007/s00705-006-0849-1.
-
28.
Spagnolo AM, Orlando P, Panatto D, Perdelli F, Cristina ML. An overview of carbapenem-resistant Klebsiella pneumoniae: epidemiology and control measures. Rev Med Microbiol. 2014;25(1):7-14. https://doi.org/10.1097/MRM.0b013e328365c51e.
-
29.
Su T, Qiu Y, Hua X, Ye B, Luo H, Liu D, et al. Novel opportunity to reverse antibiotic resistance: to explore traditional Chinese medicine with potential activity against antibiotics-resistance bacteria. Front Microbiol. 2020;11:610070. [PubMed ID: 33414777]. [PubMed Central ID: PMC7782309]. https://doi.org/10.3389/fmicb.2020.610070.
-
30.
Avershina E, Shapovalova V, Shipulin G. Fighting antibiotic resistance in hospital-acquired infections: current state and emerging technologies in disease prevention, diagnostics and therapy. Front Microbiol. 2021;12:707330. [PubMed ID: 34367112]. [PubMed Central ID: PMC8334188]. https://doi.org/10.3389/fmicb.2021.707330.
-
31.
O'Flynn G, Ross RP, Fitzgerald GF, Coffey A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl Environ Microbiol. 2004;70(6):3417-24. [PubMed ID: 15184139]. [PubMed Central ID: PMC427753]. https://doi.org/10.1128/AEM.70.6.3417-3424.2004.
-
32.
Kumari S, Harjai K, Chhibber S. Isolation and characterization of Klebsiella pneumoniae specific bacteriophages from sewage samples. Folia Microbiol (Praha). 2010;55(3):221-7. [PubMed ID: 20526833]. https://doi.org/10.1007/s12223-010-0032-7.
-
33.
Chaturongakul S, Ounjai P. Phage-host interplay: examples from tailed phages and Gram-negative bacterial pathogens. Front Microbiol. 2014;5:442. [PubMed ID: 25191318]. [PubMed Central ID: PMC4138488]. https://doi.org/10.3389/fmicb.2014.00442.
-
34.
Drulis-Kawa Z, Mackiewicz P, Kesik-Szeloch A, Maciaszczyk-Dziubinska E, Weber-Dabrowska B, Dorotkiewicz-Jach A, et al. Isolation and characterisation of KP34--a novel phiKMV-like bacteriophage for Klebsiella pneumoniae. Appl Microbiol Biotechnol. 2011;90(4):1333-45. [PubMed ID: 21327407]. [PubMed Central ID: PMC3082699]. https://doi.org/10.1007/s00253-011-3149-y.
-
35.
Kesik-Szeloch A, Drulis-Kawa Z, Weber-Dabrowska B, Kassner J, Majkowska-Skrobek G, Augustyniak D, et al. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol J. 2013;10:100. [PubMed ID: 23537199]. [PubMed Central ID: PMC3620542]. https://doi.org/10.1186/1743-422X-10-100.
-
36.
Dalmasso M, Strain R, Neve H, Franz CM, Cousin FJ, Ross RP, et al. Three New Escherichia coli Phages from the Human Gut Show Promising Potential for Phage Therapy. PLoS One. 2016;11(6). e0156773. [PubMed ID: 27280590]. [PubMed Central ID: PMC4900583]. https://doi.org/10.1371/journal.pone.0156773.
-
37.
Santos SB, Carvalho C, Azeredo J, Ferreira EC. Population dynamics of a Salmonella lytic phage and its host: Implications of the host bacterial growth rate in modelling. PLoS One. 2014;9(7). e102507. [PubMed ID: 25051248]. [PubMed Central ID: PMC4106826]. https://doi.org/10.1371/journal.pone.0102507.
-
38.
Bao H, Zhang H, Wang R. Isolation and characterization of bacteriophages of Salmonella enterica serovar Pullorum. Poult Sci. 2011;90(10):2370-7. [PubMed ID: 21934022]. https://doi.org/10.3382/ps.2011-01496.
-
39.
Kim M, Kim S, Ryu S. Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J Virol. 2012;86(19):10894. [PubMed ID: 22966187]. [PubMed Central ID: PMC3457314]. https://doi.org/10.1128/JVI.01796-12.
-
40.
Ryan EM, Gorman SP, Donnelly RF, Gilmore BF. Recent advances in bacteriophage therapy: how delivery routes, formulation, concentration and timing influence the success of phage therapy. J Pharm Pharmacol. 2011;63(10):1253-64. [PubMed ID: 21899540]. https://doi.org/10.1111/j.2042-7158.2011.01324.x.
-
41.
Wintachai P, Naknaen A, Thammaphet J, Pomwised R, Phaonakrop N, Roytrakul S, et al. Characterization of extended-spectrum-beta-lactamase producing Klebsiella pneumoniae phage KP1801 and evaluation of therapeutic efficacy in vitro and in vivo. Sci Rep. 2020;10(1):11803. [PubMed ID: 32678251]. [PubMed Central ID: PMC7367294]. https://doi.org/10.1038/s41598-020-68702-y.
-
42.
Zhang C, Yuan J, Guo C, Ge C, Wang X, Wei D, et al. Identification and complete genome of lytic "Kp34likevirus" phage vB_KpnP_Bp5 and therapeutic potency in the treatment of lethal Klebsiella pneumoniae infections in mice. Virus Res. 2021;297:198348. [PubMed ID: 33631221]. https://doi.org/10.1016/j.virusres.2021.198348.
-
43.
Wilkowske HH, Nelson FE, Parmelee CE. Heat inactivation of bacteriophage strains active against lactic streptococci. Appl Microbiol. 1954;2(5):250-3. [PubMed ID: 13208170]. [PubMed Central ID: PMC1057005]. https://doi.org/10.1128/am.2.5.250-253.1954.
-
44.
Yamaki S, Omachi T, Kawai Y, Yamazaki K. Characterization of a novel Morganella morganii bacteriophage FSP1 isolated from river water. FEMS Microbiol Lett. 2014;359(2):166-72. [PubMed ID: 25168469]. https://doi.org/10.1111/1574-6968.12560.
-
45.
Zhang J, Hong Y, Fealey M, Singh A, Walton K, Martin C, et al. Physiological and molecular characterization of salmonella bacteriophages previously used in phage therapy. J Food Protect. 2015;78(12):2143-9. https://doi.org/10.4315/0362-028x.Jfp-14-350.
-
46.
Pallavi B, Puneeth TG, Shekar M, Girisha SK. Isolation, characterization and genomic analysis of vB-AhyM-AP1, a lytic bacteriophage infecting Aeromonas hydrophila. J Appl Microbiol. 2021;131(2):695-705. [PubMed ID: 33420733]. https://doi.org/10.1111/jam.14997.
-
47.
Chen L, Yuan S, Liu Q, Mai G, Yang J, Deng D, et al. In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front Microbiol. 2018;9:1476. [PubMed ID: 30034378]. [PubMed Central ID: PMC6043867]. https://doi.org/10.3389/fmicb.2018.01476.
-
48.
Soleimani Sasani M, Eftekhar F, Hosseini M. Isolation and Characterization of a Klebsiella pneumoniae Specific Lytic Bacteriophage from a Hospital Waste-water Treatment Plant. J Med Microbiol Infect Dis. 2019;7(1):6-11. https://doi.org/10.29252/JoMMID.7.1.2.6.
-
49.
Jamal M, Hussain T, Das CR, Andleeb S. Characterization of Siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. J Med Microbiol. 2015;64(Pt 4):454-62. [PubMed ID: 25681321]. https://doi.org/10.1099/jmm.0.000040.
-
50.
Komijani M, Bouzari M, Rahimi F. Detection and characterization of a novel lytic bacteriophage (vB-KpneM-Isf48) against Klebsiella pneumoniae isolates from infected wounds carrying antibiotic-resistance genes (TEM, SHV, and CTX-M). Iran Red Crescent Med J. 2016;19(2). https://doi.org/10.5812/ircmj.34475.
-
51.
Carl G, Jackel C, Grutzke J, Hertwig S, Grobbel M, Malorny B, et al. Complete genome sequence of the temperate Klebsiella pneumoniae phage KPP5665-2. Genome Announc. 2017;5(43). [PubMed ID: 29074652]. [PubMed Central ID: PMC5658490]. https://doi.org/10.1128/genomeA.01118-17.
-
52.
Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013;8(6):769-83. [PubMed ID: 23701332]. https://doi.org/10.2217/fmb.13.47.
-
53.
Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol. 2010;11(1):2-14. [PubMed ID: 20214604]. https://doi.org/10.2174/138920110790725311.
-
54.
Bull JJ, Vegge CS, Schmerer M, Chaudhry WN, Levin BR. Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS One. 2014;9(4). e94690. [PubMed ID: 24743264]. [PubMed Central ID: PMC3990542]. https://doi.org/10.1371/journal.pone.0094690.
-
55.
Vieira A, Silva YJ, Cunha Â, Gomes NCM, Ackermann HW, Almeida A. Phage therapy to control multidrug-resistant Pseudomonas aeruginosa skin infections: in vitro and ex vivo experiments. Eur J Clin Microbiol Infect Dis. 2012;31(11):3241-9. https://doi.org/10.1007/s10096-012-1691-x.
-
56.
Kortright KE, Chan BK, Koff JL, Turner PE. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe. 2019;25(2):219-32. [PubMed ID: 30763536]. https://doi.org/10.1016/j.chom.2019.01.014.
-
57.
Torres-Barcelo C. Phage therapy faces evolutionary challenges. Viruses. 2018;10(6). [PubMed ID: 29895791]. [PubMed Central ID: PMC6024868]. https://doi.org/10.3390/v10060323.
-
58.
Ul Haq I, Chaudhry WN, Andleeb S, Qadri I. Isolation and partial characterization of a virulent bacteriophage IHQ1 specific for Aeromonas punctata from stream water. Microb Ecol. 2012;63(4):954-63. [PubMed ID: 21947462]. https://doi.org/10.1007/s00248-011-9944-2.