Abstract
Background:
The O-antigen is one of the uropathogenic Escherichia coli (UPEC) virulence factors used as a biomarker to classify E. coli strains.Objectives:
In this study, the relationship between antibiotic resistance patterns and O-serogroups was investigated in UPEC strains isolated from patients with urinary tract infections (UTIs) in southern Iraq.Methods:
A total of 100 UPEC isolates from the urine specimens of patients with UTIs within the age range of 4 months to 78 years in various southern Iraqi hospitals were collected (May 2017 to January 2018) and confirmed using biochemical tests (e.g., Analytical Profile Index 20E). Antibiotic susceptibility tests were performed using the disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines. The multiple polymerase chain reaction technique was applied to investigate the prevalence of O-serogroups.Results:
The most frequent serogroups in the E. coli isolates were O8 (27.7%) and O25 (24.4%); however, serogroup O83 was not observed in the samples. Serogroups O75, O6, O16, and O18 had the lowest frequency (1.1%) among the examined isolates. Furthermore, 10% of the isolates did not belong to any of the examined serogroups. The phenotypic tests showed that the highest and the lowest resistance belonged to piperacillin (92%) and imipenem (5%), respectively. Serogroups O4 and O21 showed the highest drug resistance; nevertheless, serogroups O75, O18, and O1 showed the lowest drug resistance. Additionally, 94% of the isolates were resistant to three or more classes of antibiotics.Conclusions:
According to the results, UPEC isolates showed high resistance to common antibiotics; however, they were sensitive to imipenem and amikacin. Serogroups O8 and O25 were the most common among UPEC isolates. Moreover, O4 and O21 showed the highest drug resistance. There was a direct relationship between antimicrobial resistance and O-serogroups in UPEC isolates.Keywords
Urinary Tract Infection Antibiotic Resistance O-antigen Uropathogenic Escherichia coli
1. Background
The urinary tract infection (UTI), mainly caused by Escherichia coli, is the most common bacterial infection in humans after respiratory tract infections (1, 2). Escherichia coli is the cause of 80 - 90% of community-acquired UTIs and 30 - 50% of hospital-acquired UTIs. The UTIs are among the main causes of hospitalization; such infections are accompanied by significant complications and high health care costs. Each year, about 150 million individuals are diagnosed with UTIs, and more than 6 billion dollars are spent on UTI treatment worldwide (3). The urinary system structures vary with male and female anatomy; accordingly, the prevalence and frequency of UTIs in female patients are much higher than in male patients, and 50 - 60% of adult women experience UTIs at least once in their lifetime (4).
Escherichia coli includes a variety of pathogenic and commensal strains typically identified by serotyping their surface antigens, including the O-antigen (Lipopolysaccharides), the H-antigen (Flagellate), and in some cases, the K-antigen (capsule) (5). Among the somatic antigens, the O-antigen, which is made up of repeated oligosaccharide units, is part of the lipopolysaccharide in the outer membrane of Gram-negative bacteria and is more important than others. The O-antigen is considered a biomarker used to classify and serogroup E. coli strains. To date, more than 181 O-antigen groups have been identified for E. coli; among the identified O-antigens, O83, O75, O25, O22, O21, O18, O16, O15, O8, O7, O6, O4, O2, and O1 serogroups are involved in UTIs (6). Each of the aforementioned serogroups plays an essential role in the clinical evidence of UTIs, and the prevalence of each serogroup varies in different regions (7).
One of the most common problems in medical centers is the increasing prevalence of microbial resistance. The UTIs are often treated with broad-spectrum antibiotics; therefore, the identification of resistance patterns is necessary to improve experimental antibiotic treatment strategies (8). The degree of drug resistance varies in different geographical regions and is higher in areas where antibiotics are used experimentally without being controlled (9). Antibiotic resistance limits treatment and increases treatment costs and duration. Therefore, physicians’ knowledge of the antibiotic resistance profile of a specific geographical area’s population can be considerably helpful for the treatment of UTIs.
2. Objectives
Despite the high prevalence of UTIs in Iraq, few studies have been conducted to determine the antibiotic resistance profile and the frequency of O-serogroups in uropathogenic Escherichia coli (UPEC) in the Iraqi population. Therefore, the present study was conducted to determine the degree of drug resistance and the prevalence of O-serogroups in UPEC strains isolated from patients with UTIs and investigate the relationship between O-serogroups and antibiotic resistance patterns among UTI patients referring to different hospitals in the southern part of Iraq.
3. Methods
3.1. Collection and Identification of Bacterial Strains
In the present study, 385 urine samples from patients with UTI symptoms, within the age range of 4 months to 78 years, referring to Maysan Governorate (Iraq) hospitals within May 2017 to January 2018, were collected and examined. The sampling was performed according to the McGeer criteria (10). Urine cultures yielded to colony-forming units (CFU) of greater than 100,000 CFU per milliliter were considered UTIs. A total of 100 UPEC bacteria were isolated, purified, identified using the Analytical Profile Index 20E kit (bioMérieux, France), and stored at -20°C until use.
3.2. Antimicrobial Susceptibility
The antibiogram testing was performed using the disk diffusion method on Mueller-Hinton agar (Merck, Germany) according to the Clinical and Laboratory Standards Institute (2020) guidelines (11). Commercial antibiotic disks (Bioanalyse, Turkey), including ticarcillin (75 μg), amoxicillin-clavulanic acid (20/10 μg), piperacillin (100 μg), trimethoprim-sulfamethoxazole (1.25/23.75 μg), doxycycline (30 μg), ceftazidime (30 μg), cefixime (5 μg), ceftriaxone (10 μg), nitrofurantoin (300 μg), cefoxitin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), norfloxacin (10 μg), amikacin (30 μg), and imipenem (10 μg), were used in this study. Escherichia coli ATCC 25922 strain was used as a control in the assay.
3.3. Identification of Serotyping Groups
The multiplex polymerase chain reaction (PCR) technique was applied to evaluate the prevalence of serotyping groups among patients with UTIs. Deoxyribonucleic acid (DNA) extraction was performed via boiling, and serotyping was performed in four separate groups (i.e., A, B, C, and D). Table 1 shows the sequences of primers used for each gene. Each PCR was carried out in a 25 μL volume containing Master Mix RED (Ampliqon, Denmark), 9 μL for groups A and B and 10 μL for groups C and D [each primer (F&R) 1 μL (10 pmol), DNA template 2 μL, and the rest distilled water until 25 μL]. The PCR (Bio-Rad, Germany) amplifications were performed as follows:
Sequences of Used Primers, the Amount of Master Mix Used in Each Run, and the Temperature Program of Polymerase Chain Reaction for Each Primer Set
| Serogroups Primers and Sequence (5'-3') | Size of Product (bp) | Reference |
|---|---|---|
| Group A | ||
| O1 | 1098 | (11) |
| F:GTGAGCAAAAGTGAAATAAGGAACG | ||
| R: CGCTGATACGAATACCATCCTAC | ||
| O2 | 770 | (11) |
| F: AGTGAGTTACTTTTTAGCGATGGAC | ||
| R:AGTTTAGTATGCCCCTGACTTTGAA | ||
| O7 | 610 | (11) |
| F: CTATCAAAATACCTCTGCTGGAATC | ||
| R: TGGCTTCGAGATTAAACCTATTCCT | ||
| O8 | 448 | (12) |
| F: CCAGAGGCATAATCAGAAATAACAG | ||
| R: GCAGAGTTAGTCAACAAAAGGTCAG | ||
| Group B | ||
| O6 | 783 | (11) |
| F: GGATGACGATGTGATTTTGGCTAAC | ||
| R: TCTGGGTTTGCTGTGTATGAGGC | ||
| O16 | 302 | (12) |
| F: GGTTTCAATCTCACAGCAACTCAG | ||
| R: GTTAGAGGGATAATAGCCAAGCGG | ||
| O75 | 511 | (11) |
| F: GAGATATACATGGGGAGGTAGGCT | ||
| R: ACCCGATAATCATATTCTTCCCAAC | ||
| O18 | 551 | (11) |
| F: GTTCGGTGGTTGGATTACAGTTAG | ||
| R: CTACTATCATCCTCACTGACCACG | ||
| Group C | ||
| O22 | 468 | (11) |
| F: TTCATTGTCGCCACTACTTTCCG | ||
| R: GAAACAGCCCATGACATTACTACG | ||
| O25 | 230 | (11) |
| F: AGAGATCCGTCTTTTATTTGTTCGC | ||
| R: GTTCTGGATACCTAACGCAATACCC | ||
| O83 | 362 | (12) |
| F: GTACACCAGGCAAACCTCGAAAG | ||
| R: TTCTGTAAGCTAATGAATAGGCACC | ||
| Group D | ||
| O4 | 664 | (11) |
| F: TTGTTGCGATAATGTGCATGTTCC | ||
| R: AATAATTTGCTATACCCACACCCTC | ||
| O15 | 183 | (12) |
| F: TCTTGTTAGAGTCATTGGTGTATCG | ||
| R: ATAAAACGAGCAAGCACCACACC | ||
| O21 | 209 | (11) |
| F: CTGCTGATGTCGCTATTATTGCTG | ||
| R:TGAAAAAAAGGGAAACAGAAGAGCC | ||
Initial denaturation at 95°C for 5 minutes; 30 cycles of denaturation at 95°C for 30 seconds, annealing at 50°C for 60 seconds, and extension at 72°C for 60 seconds; a final extension at 72°C for 5 minutes (for groups A and B)
Initial denaturation at 95°C for 5 minutes; 30 cycles of denaturation at 95°C for 60 seconds, annealing at 50°C for 60 seconds, and extension at 72°C for 90 seconds; a final extension at 72°C for 10 minutes (for groups C and D)
3.4. Data Analysis
The collected data were analyzed using SPSS statistical package (version 24) to investigate the relationship between antibiotic resistance patterns and UPEC serogroups isolated from patients with UTIs. The descriptive analysis of parametric and non-parametric variables was performed using the chi-square and Fisher’s exact tests. A P-value of less than 0.05 was considered statistically significant.
4. Results
4.1. Population Study
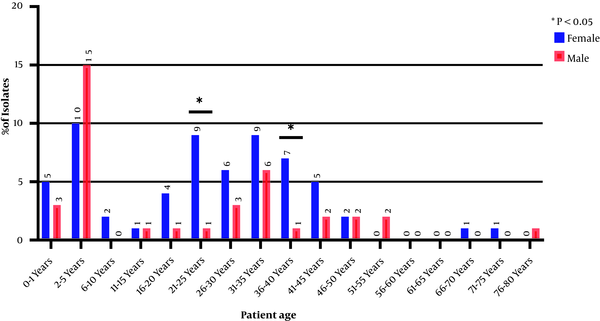
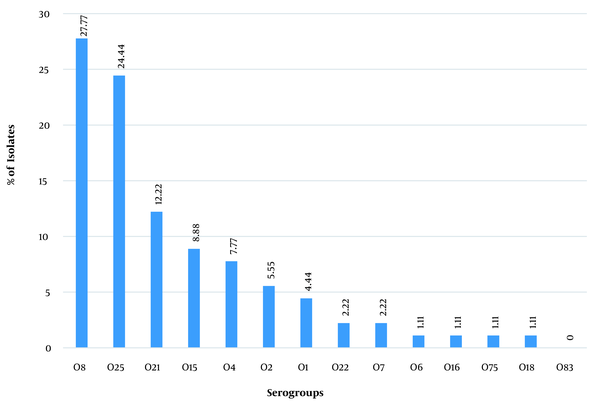
A total of 100 E. coli isolates were examined in the present study. According to the results, 62 and 38% of the urine samples belonged to female and male patients, respectively. Accordingly, a significant relationship was observed between the incidence of UTI and gender (P < 0.05). The collected strains were mostly related to the age group of 2 - 5 years, and the percentage of the samples isolated from the age group of > 55 years showed a significant decrease (Figure 1). The O-serotyping results (Figure 2) indicated that 90% of the examined strains were positive for one of the O-antigens; serogroups O8 (27.7%), O25 (24.4%), and O21 (12.2%) had the highest frequencies, and serogroup O83 was not noticed in any samples (Figure 3).
Distribution of uropathogenic Escherichia coli isolates based on patients’ age and gender
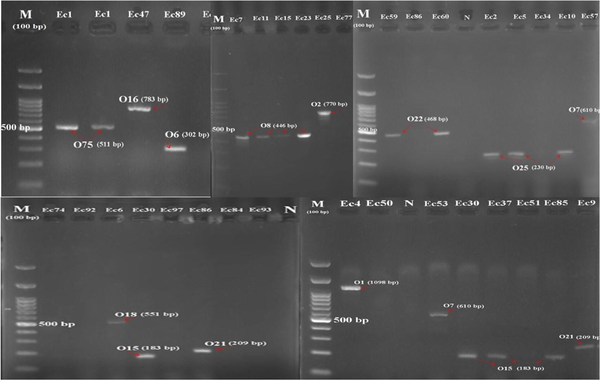
Results of multiplex polymerase chain reaction specific for serogroups (M, deoxyribonucleic acid ladder 100 bp; N, negative control; and Ec1-ec93, different serogroups).
Frequency of O-serogroups among uropathogenic Escherichia coli isolates
4.2. Antibiotic Resistance Profile
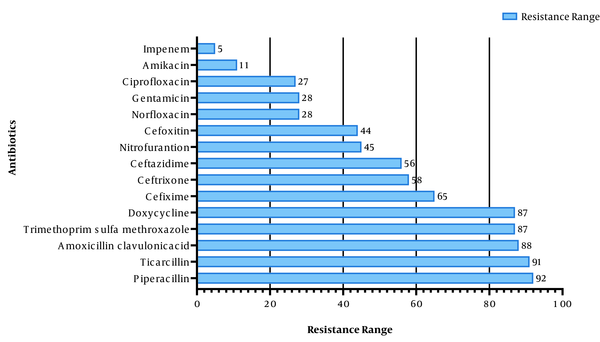
Among the studied UPEC isolates in this study, the highest resistance was observed against piperacillin (92%), ticarcillin (91%), and amoxicillin-clavulanic acid (88%). Amikacin (11%) and imipenem (5%) showed the highest susceptibility (Figure 4), and 94% of the strains were resistant to three or more classes of antibiotics [i.e., multidrug resistance (MDR)]. Statistical results showed that the overall resistance of the tested antibiotics was higher in male patients than in female ones (P < 0.038). There was also a significant relationship between antibiotic resistance and age; accordingly, antibiotic resistance was higher in older patients (P < 0.05).
Drug resistance profile among uropathogenic Escherichia coli isolates
4.3. Frequency of Serogroups by Age and Gender
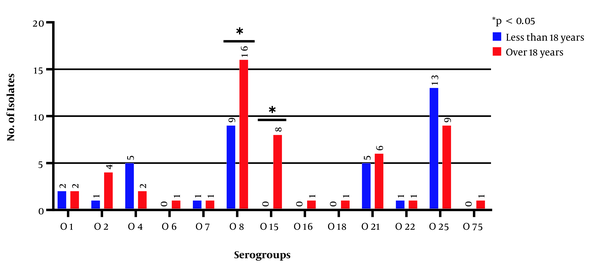
The investigation of the frequency of serogroups based on patients’ gender showed that 11 serogroups existed in the UPEC strains isolated from male patients; the three serogroups of O75, O83, and O16 did not exist in those strains; three isolates were non-typeable, and the serogroups O8 and O25 (each with 10 isolates) had the highest frequencies. In the UPEC strains isolated from female patients, 11 serogroups were typable, and 7 isolates were non-typeable; the serogroups O83, O18, and O6 did not exist in any of the strains, and the serogroups O8 (15 isolates) and O25 (12 isolates) showed the highest frequencies. The results revealed that out of 90 serogroup-positive samples, 48 and 42 samples were related to the age ranges of > 18 and < 18 years, respectively. The serogroups O6, O15, O16, O18, and O75 were not observed within the age range of < 18 years. The frequencies of O25 and O4 serogroups’ isolates were higher within the age range of < 18 years than the age range of > 18 years, and the frequencies of serogroups O22, O1, and O7 were equal in both age groups. The frequencies of serogroup-negative samples were equal (n = 5) in both age groups (Figure 5), and the frequencies of serogroups were significant between the two age groups (P < 0.05).
Frequency of serogroups based on the age of patients with urinary tract infections
4.4. Relationship Between O-Serogroups and Drug Resistance
Based on the results, the isolates of serogroups O4 and O21 showed the highest drug resistance; however, the isolates of serogroups O75, O18, and O1 showed the lowest resistance. Among different serogroups, the highest and lowest resistance was observed to amoxicillin-clavulanic acid and imipenem, respectively (Table 2). The pattern of antibiotic resistance variation showed that all the strains related to serogroups O15, O8, and O25 were resistant to at least one of the 16 tested antibiotics; in comparison, the strains of serogroup O75 had the lowest antibiotic resistance variation (only to five antibiotics). Imipenem-resistant isolates belonged to O2, O8, O15, and O25 serogroups.
Frequency of Drug Susceptibility of Isolates Among O Serotype Groups
| Serotypes | O1 | O2 | O4 | O6 | O7 | O8 | O15 | O16 | O18 | O21 | O22 | O25 | O75 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample numbers | 4 | 5 | 7 | 1 | 2 | 25 | 8 | 1 | 1 | 11 | 2 | 22 | 1 | 90 |
| Amikacin | - | - | - | - | - | 1 | 1 | - | - | 3 | - | 4 | - | 9 |
| Ceftazidime | 2 | 2 | 5 | 1 | 1 | 12 | 5 | - | - | 10 | 2 | 12 | - | 52 |
| Gentamicin | - | - | 2 | - | - | 6 | 2 | - | 1 | 5 | - | 8 | - | 24 |
| Ciprofloxacin | - | - | 2 | - | 1 | 9 | 4 | - | - | 6 | - | 3 | - | 25 |
| Norfloxacin | 1 | 1 | 2 | - | 1 | 10 | 4 | 1 | - | 7 | - | 6 | - | 33 |
| Nitrofurantoin | 1 | 5 | 3 | 1 | 1 | 16 | 4 | - | - | 7 | 1 | 11 | - | 50 |
| Cefixime | 2 | 4 | 5 | 1 | 1 | 12 | 6 | - | 1 | 11 | 1 | 13 | - | 57 |
| Imipenem | - | 1 | - | - | - | 2 | 1 | - | - | - | - | 3 | - | 7 |
| Cefoxitin | 1 | - | 5 | - | - | 11 | 4 | - | - | 8 | 1 | 10 | - | 40 |
| Ceftriaxone | 2 | 1 | 25 | - | 1 | 12 | 5 | 1 | 1 | 10 | 1 | 14 | 1 | 74 |
| Amoxicillin/ clavulanic acid | 4 | 5 | 7 | 1 | 2 | 24 | 8 | 1 | - | 11 | 2 | 21 | 1 | 87 |
| Piperacillin | 4 | 5 | 7 | 1 | 2 | 23 | 7 | 1 | 1 | 9 | 2 | 20 | 1 | 83 |
| Ticarcillin | 3 | 3 | 7 | 1 | 1 | 23 | 8 | 1 | 1 | 11 | 2 | 17 | 1 | 79 |
| Trimethoprim/ sulfamethoxazole | 2 | 4 | 6 | 1 | 2 | 25 | 8 | 1 | - | 11 | 2 | 18 | - | 80 |
| Doxycycline | 4 | 5 | 7 | 1 | 2 | 20 | 8 | 1 | 1 | 9 | 2 | 19 | 1 | 80 |
| Percentage (%) | 40 | 48 | 79 | 53.3 | 50 | 54.9 | 62.5 | 46.6 | 40 | 71.5 | 53.3 | 54.2 | 33.3 | - |
5. Discussion
The present study has been the first report on the relationship between the serogroups of UPEC, causing UTIs, and drug resistance profiles in Southern Iraq. According to the results, the highest number of collected strains belonged to the age range of 1 - 5 years. In addition, 90% of the 100 samples had the desired serogroups. Moreover, 10% of the samples were not related to any of the O-serogroups, indicating that the serogroups used in this study were the most frequent and the most important among isolates causing UTIs. Serogroups O8 (27.7%) and O25 (24.4%) were the most frequent in the examined UPEC strains. The results of this study showed a significant relationship between serogroups O8 and O25 with age; accordingly, O8 and O25 serogroups showed high frequency within the age ranges of > 18 and < 18 years, respectively. Serogroup O83 was not detected in any of the examined UPEC isolates.
Serogroups O8, O15, O21, and O25 showed the highest frequencies; this finding is in line with the findings of previous studies in Iran and Mexico (13, 14). Consequently, it can be concluded that the aforementioned four serogroups are highly important in the development of UTIs by E. coli. Goudarzi et al. evaluated the O-serogroups of E. coli strains isolated from Iranian patients with UTIs and showed that out of 150 E. coli strains, the highest frequencies were related to serogroups O16 (8%), O21 (4%), O8 (3.5%), and O1 (3.13%). Goudarzi et al. also reported that serogroups O83 and O22 were not present in any of the isolates (15). In another study, Wabale et al. examined the prevalence of serogroups among UPEC strains isolated from the patients in one of Mumbai’s hospitals in India. They reported that all serogroups were available in their examined 105 UPEC strains, and serogroups O25 and O120 had the highest frequencies (16). The results of the aforementioned study concerning the prevalence of serogroup O25 are consistent with the results of the present study.
In this study, the highest frequencies were related to O8, O15, O21, and O25 serogroups. Moreover, O21 and O15 serogroups had the highest drug resistance; nonetheless, serogroups O75 and O18 showed the lowest drug resistance. The results of the present study did not show any correlation or association between the frequency of serogroups and drug resistance, which is in line with some results obtained by researchers, such as Sadeghi et al. However, some researchers could find a relationship between some serogroups and drug resistance (13, 17). In different parts of the world, the resistance of E. coli to the penicillin class of antibiotics is high and even increasing day by day; accordingly, in some studies, the level of resistance against the penicillin class of antibiotics has been reported as 100%, which is consistent with the present study’s findings (18-20).
In the present study, piperacillin (92%) had the lowest drug activity, with moderate resistance, compared to the cephalosporin class of antibiotics. These findings are in line with the results of previous studies (21, 22). In general, the penicillins and cephalosporins are not active against UTIs in developing countries, such as Iraq; therefore, according to the results, such antibiotics should not be used in the treatment of UTIs. In recent years, quinolones, especially ciprofloxacin, have been used to treat E. coli infections, and studies in different parts of the world have shown that quinolones are still active against UTIs (23, 24). However, 27% of the UPEC isolates in the present study showed resistance to ciprofloxacin.
Recently, carbapenem antibiotics have been recognized as effective treatments for antibiotic-resistant UTIs, and this class of antibiotics is now widely used in the treatment of complex microbial infections (25). In the current study, amikacin (11%) and imipenem (5%) showed the highest drug activity against uropathogenic isolates, which was in line with the results of other studies conducted in Iran, Pakistan, and Mexico (21, 22, 26). Other studies have reported that amikacin might be a good option for the treatment of UTIs caused by E. coli (27, 28). Currently, amikacin and imipenem are among the most suitable treatments for UTIs in numerous countries.
At present, the emergence of MDR-UPEC strains has led to treatment failures, turning UTIs into serious health problems (29). In this regard, 94% of the isolates examined in this study were resistant to multiple antibiotic classes, which indicated a high level of MDR in southern Iraq. In an Iranian study, Dehbanipour et al. reported that the degrees of multiple antibiotic resistance in inpatients and outpatients were 68 and 61%, respectively (30). The aforementioned results indicated that the degrees of resistance to various antibiotics were higher in Iraq than in the neighboring countries, which could be due to inappropriate and excessive use of antibiotics over the past years.
Antibiotic resistance has increased due to some factors, such as frequent and irrational administration of antibiotics, mutations, and transmission of resistance agents. By the comparison of different patterns of drug resistance, it can be concluded that there are different therapeutic responses to antimicrobial drugs worldwide or even in one country. Consequently, regular and frequent research on antibiotic resistance should be carried out to control the distribution of antibiotic resistance. Furthermore, the high prevalence of O8 and O25 serogroups suggests a close relationship between serogroups and characteristic features of UPEC isolates. However, it should be noted that there were some limitations in this study regarding the sampling, such as sampling from a few hospitals and incorrect sampling by some patients, which might affect the results.
5.1. Conclusions
This study showed the relationship between the prevalence of O-antigen serogroups and the pattern of antibiotic resistance in the UPEC strains isolated from the UTI patients in southern Iraq. Serogroups O8 and O25 had the highest frequencies in the examined isolates. The relationship between O-serogroups and antibiotic resistance among the UPEC strains isolated from Iraqi patients indicated that isolates of serogroups O75 and O18 had the lowest degrees of drug resistance. In contrast, the isolates of O4 and O21 serogroups had the highest degrees of drug resistance. The E. coli isolates showed high degrees of resistance to commonly used antibiotics for the treatment of UTIs; nevertheless, the E. coli isolates were sensitive to imipenem and amikacin. The results of this study enhanced the understanding of the importance of these serogroups in the pathogenicity of UPEC isolates, which can be helpful to develop treatments and prevention strategies for UTIs.
Acknowledgements
References
-
1.
Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013;12:8. [PubMed ID: 23627669]. [PubMed Central ID: PMC3651382]. https://doi.org/10.1186/1476-0711-12-8.
-
2.
Bakhtiari S, Mahmoudi H, Seftjani SK, Amirzargar MA, Ghiasvand S, Ghaffari ME, et al. Antibiotic resistance pattern and phylogenetic groups of the uropathogenic Escherichia coli isolates from urinary tract infections in Hamedan, west of Iran. Iran J Microbiol. 2020;12(5):388-94. [PubMed ID: 33603992]. [PubMed Central ID: PMC7867701]. https://doi.org/10.18502/ijm.v12i5.4598.
-
3.
Zhao L, Gao S, Huan H, Xu X, Zhu X, Yang W, et al. Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiology (Reading). 2009;155(Pt 5):1634-44. [PubMed ID: 19372154]. https://doi.org/10.1099/mic.0.024869-0.
-
4.
Medina M, Castillo-Pino E. An introduction to the epidemiology and burden of urinary tract infections. Ther Adv Urol. 2019;11:1756287219832170. [PubMed ID: 31105774]. [PubMed Central ID: PMC6502976]. https://doi.org/10.1177/1756287219832172.
-
5.
Li D, Liu B, Chen M, Guo D, Guo X, Liu F, et al. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods. 2010;82(1):71-7. [PubMed ID: 20434495]. https://doi.org/10.1016/j.mimet.2010.04.008.
-
6.
Liu B, Furevi A, Perepelov AV, Guo X, Cao H, Wang Q, et al. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev. 2020;44(6):655-83. [PubMed ID: 31778182]. [PubMed Central ID: PMC7685785]. https://doi.org/10.1093/femsre/fuz028.
-
7.
Sharma S, Kaur N, Malhotra S, Madan P, Ahmad W, Hans C. Serotyping and antimicrobial susceptibility pattern of Escherichia coli isolates from urinary tract infections in pediatric population in a tertiary care hospital. J Pathog. 2016;2016:2548517. [PubMed ID: 27047691]. [PubMed Central ID: PMC4800102]. https://doi.org/10.1155/2016/2548517.
-
8.
Martins A, Hunyadi A, Amaral L. Mechanisms of resistance in bacteria: An evolutionary approach. Open Microbiol J. 2013;7:53-8. [PubMed ID: 23560029]. [PubMed Central ID: PMC3613773]. https://doi.org/10.2174/1874285801307010053.
-
9.
Datta P, Gupta V, Sidhu S. Extended spectrum beta lactamase positive uropathogenic E. coli epidemiological factors and resistance. BJMP. 2014;7(2). a718.
-
10.
Rowe TA, Juthani-Mehta M. Urinary tract infection in older adults. Aging health. 2013;9(5). [PubMed ID: 24391677]. [PubMed Central ID: PMC3878051]. https://doi.org/10.2217/ahe.13.38.
-
11.
Clinical and Laboratory Standards Institute. CLSI supplement M100. Performance standards for antimicrobial susceptibility testing. 30th ed. Wayne, USA: Clinical and Laboratory Standards Institute; 2020.
-
12.
Tajbakhsh E, Ahmadi P, Abedpour-Dehkordi E, Arbab-Soleimani N, Khamesipour F. Biofilm formation, antimicrobial susceptibility, serogroups and virulence genes of uropathogenic E. coli isolated from clinical samples in Iran. Antimicrob Resist Infect Control. 2016;5:11. [PubMed ID: 27042294]. [PubMed Central ID: PMC4818419]. https://doi.org/10.1186/s13756-016-0109-4.
-
13.
Noie Oskouie A, Hasani A, Ahangarzadeh Rezaee M, Soroush Bar Haghi MH, Hasani A, Soltani E. A relationship between O-serotype, antibiotic susceptibility and biofilm formation in uropathogenic Escherichia coli. Microb Drug Resist. 2019;25(6):951-8. [PubMed ID: 30817229]. https://doi.org/10.1089/mdr.2018.0330.
-
14.
Paniagua-Contreras GL, Monroy-Perez E, Rodriguez-Moctezuma JR, Dominguez-Trejo P, Vaca-Paniagua F, Vaca S. Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. J Microbiol Immunol Infect. 2017;50(4):478-85. [PubMed ID: 26433755]. https://doi.org/10.1016/j.jmii.2015.08.005.
-
15.
Goudarzi V, Mirzaee M, Ranjbar R. O-serogrouping of Escherichia coli strains isolated from patients with Urinary tract infection by using PCR method. Iran J Med Microbiol. 2017;10(6):1-8.
-
16.
Wabale VR, Bharadwaj RS, Joshi AA, Chowdhary AS. Serotypes, hemolysin production and drug resistance among uropathogenic Escherichia coli isolated at a tertiary care hospital in Mumbai. Int J Curr Res. 2015;7(7):18596-600.
-
17.
Sadeghi A, Halaji M, Fayyazi A, Havaei SA. Characterization of plasmid-mediated quinolone resistance and serogroup distributions of uropathogenic Escherichia coli among Iranian kidney transplant patients. Biomed Res Int. 2020;2020:2850183. [PubMed ID: 33195692]. [PubMed Central ID: PMC7641683]. https://doi.org/10.1155/2020/2850183.
-
18.
Olowe OA, Eniola KIT, Olowe RA, Olayemi AB. Starch paper technique is easy to detect beta lactamase detection from cases of diarrheagenic Escherichia coli in Osogbo. Life Sci J. 2007;4:72-4.
-
19.
Sabir S, Anjum AA, Ijaz T, Ali MA, Rehman MU, Nawaz M. Isolation and antibiotic susceptibility of E. coli from urinary tract infections in a tertiary care hospital. Pak J Med Sci. 1969;30(2). https://doi.org/10.12669/pjms.302.4289.
-
20.
Rashki A, Rahdar M, Rashki Ghalehnoo Z. Characterization of uropathogenic Escherichia coli: Distribution of adhesin-encoding genes and O-serotypes among ciprofloxacin susceptible and resistant isolates. Jundishapur J Microbiol. 2019;12(9). e89179. https://doi.org/10.5812/jjm.89179.
-
21.
Salehzadeh A, Zamani H. Characterization of (uropathogenic) E. coli isolated from urinary tract infections: phylogenetic typing and distribution of virulence-associated traits. Br J Biomed Sci. 2018;75(1):40-2. [PubMed ID: 28709389]. https://doi.org/10.1080/09674845.2017.1336834.
-
22.
Ahmed N, Zeshan B, Naveed M, Afzal M, Mohamed M. Antibiotic resistance profile in relation to virulence genes fimH, hlyA and usp of uropathogenic E. coli isolates in Lahore, Pakistan. Trop Biomed. 2019;36(2):559-68.
-
23.
Cao D, Shen Y, Huang Y, Chen B, Chen Z, Ai J, et al. Levofloxacin versus ciprofloxacin in the treatment of urinary tract infections: Evidence-based analysis. Front Pharmacol. 2021;12:658095. [PubMed ID: 33897441]. [PubMed Central ID: PMC8060646]. https://doi.org/10.3389/fphar.2021.658095.
-
24.
Mohamed MA, Abdifetah O, Hussein FA, Karie SA. Antibiotic resistance pattern of Escherichia coli isolates from outpatients with urinary tract infections in Somalia. J Infect Dev Ctries. 2020;14(3):284-9. [PubMed ID: 32235089]. https://doi.org/10.3855/jidc.12189.
-
25.
Kot B, Gruzewska A, Szweda P, Wicha J, Parulska U. Antibiotic resistance of uropathogens isolated from patients hospitalized in district hospital in central Poland in 2020. Antibiotics (Basel). 2021;10(4). [PubMed ID: 33923389]. [PubMed Central ID: PMC8071495]. https://doi.org/10.3390/antibiotics10040447.
-
26.
Lopez-Banda DA, Carrillo-Casas EM, Leyva-Leyva M, Orozco-Hoyuela G, Manjarrez-Hernandez AH, Arroyo-Escalante S, et al. Identification of virulence factors genes in Escherichia coli isolates from women with urinary tract infection in Mexico. Biomed Res Int. 2014;2014:959206. [PubMed ID: 24895634]. [PubMed Central ID: PMC4026957]. https://doi.org/10.1155/2014/959206.
-
27.
Cho SY, Choi SM, Park SH, Lee DG, Choi JH, Yoo JH. Amikacin therapy for urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli. Korean J Intern Med. 2016;31(1):156-61. [PubMed ID: 26767869]. [PubMed Central ID: PMC4712420]. https://doi.org/10.3904/kjim.2016.31.1.156.
-
28.
Polat M, Kara SS. Once-daily intramuscular amikacin for outpatient treatment of lower urinary tract infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in children. Infect Drug Resist. 2017;10:393-9. [PubMed ID: 29138582]. [PubMed Central ID: PMC5674974]. https://doi.org/10.2147/IDR.S148703.
-
29.
Emamghorashi F, Farshad S, Kalani M. Relationship between o serotype and virulent genes in Escherichia coli causing urinary tract infections. Iran J Kidney Dis. 2011:234-7.
-
30.
Dehbanipour R, Rastaghi S, Sedighi M, Maleki N, Faghri J. High prevalence of multidrug-resistance uropathogenic Escherichia coli strains, Isfahan, Iran. J Nat Sci Biol Med. 2016;7(1):22-6. [PubMed ID: 27003964]. [PubMed Central ID: PMC4780161]. https://doi.org/10.4103/0976-9668.175020.