Abstract
Background:
Indonesia is one of the five countries with the highest number of diphtheria cases worldwide. Diphtheria is caused by the toxigenic strains Corynebacterium diphtheriae, C. ulcerans, and C. pseudotuberculosis. The diphtheria-causing bacteria can be identified using conventional and molecular methods, including polymerase chain reaction (PCR) assay. We used the PCR assay as additional testing, because in island countries like Indonesia, specimen transport is a serious challenge to maintaining bacterial survival.Objectives:
This study aimed to evaluate the PCR assay as additional testing to identify diphtheria-causing bacteria in the diphtheria laboratory.Methods:
In this cross-sectional study, a total of 178 pairs of the throat and nasal swabs from diphtheria suspected cases and close contacts were collected from seven provinces in Indonesia in 2016. All samples were directly identified by the conventional method and multiplex PCR assay. Statistical analysis was conducted using the 2 × 2 tables to determine the sensitivity and specificity of both methods, while the χ2 test was used to examine the correlation between specimen examination delay and the differentiation of results. A P-value < 0.05 was considered as statistically significant.Results:
Out of 178 examined samples, eight samples were identified as diphtheria-positive by both the conventional method and PCR assay, while nine samples were only detected by the PCR assay. All diphtheria-causing bacteria found in the positive samples were toxigenic C. diphtheriae. The diphtheria-causing bacteria were found in 27.6% of cases and 6.0% of close contacts using the PCR assay versus 13.8% of cases and 2.7% of close contacts using the conventional method. Statistical analysis showed that the PCR assay is about twice more sensitive than the conventional method. There was a significant correlation between the differentiation of results and > 72 hours’ specimen examination delay with a P-value of 0.04 (< 0.05).Conclusions:
The PCR assay is more sensitive than the conventional method to identify diphtheria-causing bacteria and may be applied as additional testing to increase the positivity rate of diphtheria-confirmed cases in Indonesia as an archipelago country where geographical factors and specimen transport are real obstacles.Keywords
1. Background
The World Health Organization (WHO) data show Indonesia as one of the countries with large diphtheria cases in recent years. India is almost in the first rank of countries with the largest diphtheria cases every year. In 2014, Nepal took second place while Indonesia descended to third place. In 2015 and 2016, Indonesia settled in the third place, whereas Madagascar replaced Nepal. In 2017, Indonesia was the second country with the highest cases globally, whereas it became the fourth country in 2018, as the cases increased in Nigeria and Yemen. Diphtheria occurred in most of Indonesia’s provinces, from the west end (Aceh) to the east end (Papua) (1, 2).
The identification of diphtheria-causing bacteria can be done by conventional or molecular methods. The conventional method is the gold standard of diphtheria laboratory examination and is used to confirm diphtheria cases. It includes isolation, purification, biochemical, and toxigenicity tests. The conventional method has some limitations. The laboratory technicians’ expertise and experience are absolute requirements because they determine the examination accuracy. It also requires more time (3 - 5 days) to obtain the result. Additionally, the conventional method needs the bacterial viability that is influenced by the antibiotic administration history, transport medium, and the time elapsed from sample collection to processing (3, 4).
Molecular methods, especially polymerase chain reaction (PCR), are the common assays for identifying diphtheria-causing bacteria. The PCR assay can be performed for clinical samples or isolates obtained from the culture (5, 6). The benefits of the PCR assay over the conventional method include faster examination (only within hours), more sensitivity, and obtained results that are relatively less affected by the duration of sample transportation and the history of antibiotic use. Since Indonesia is a large archipelago country, sample transportation is an essential matter. However, to date, the WHO has not recommended PCR as a confirmatory method for diphtheria. The WHO recommends PCR as the screening method for bacterial toxigenicity that should be confirmed by the Elek test (4, 7-9). PCR cannot be used to identify bacterial toxigenicity of non-toxigenic tox gene-bearing (NTTB) type correctly. On the other hand, we developed a PCR assay that could predict some NTTB types (10).
2. Objectives
This study aimed to evaluate the implementation of the PCR assay as additional testing to identify diphtheria-causing bacteria in the diphtheria laboratory.
3. Methods
3.1. Samples
The study was conducted from June to December 2016 with a cross-sectional design. The sample included 178 pairs of the throat and nasal swabs collected from diphtheria suspected cases and close contacts in several provinces of Indonesia, including Banten, West Borneo, East Borneo, South Sumatra, Bangka Belitung (Babel), Bali, and East Java. The samples were then transported using Amies medium at 2 - 8°C to the Prof. Dr. Sri Oemijati Research Laboratory for Infectious Diseases, Jakarta, Indonesia. The reference strains of Corynebacterium diphtheriae (NCTC 10648, NCTC 3984, and NCTC 10356), C. ulcerans (NCTC 12077), and synthetic dtxR gene of C. pseudotuberculosis were used as positive controls.
3.2. Conventional Method
The conventional method recommended by the WHO, with a minor modification, was used in this study. The samples were cultured on the cystine tellurite blood agar (CTBA) selective medium and blood agar + fosfomycin semi-selective medium, incubated at 37°C for 24 - 48 hours. The Albert staining was used to differentiate coryneform from others. The identification of diphtheria-causing bacteria was performed by API Coryne (bioMérieux, France), while bacterial toxigenicity was identified by the modified Elek test (11).
3.3. PCR Assay
The DNA extraction was performed using QIAamp DNA Minikit (QIAGEN, Germany) according to the manufacturer’s procedure. The PCR assay to identify bacterial species and toxigenicity was conducted by in-house multiplex PCR (10). The PCR results were determined based on the multiple marker bands interpretation, including 100 bp and 538 bp as toxigenicity markers (tox gene) and 162 bp, 259 bp, and 375 bp as species markers. If the 538 bp band was not accompanied by 100 bp, the result was predicted as an NTTB type.
3.4. Statistical Analysis
The 2 × 2 tables were applied as the statistical analysis to determine the sensitivity and specificity of both PCR assay and conventional method, while the χ2 test was used to correlate between specimen examination delay and the differentiation of results. The P-value of < 0.05 was considered statistically significant.
4. Results
4.1. Sampling Locations
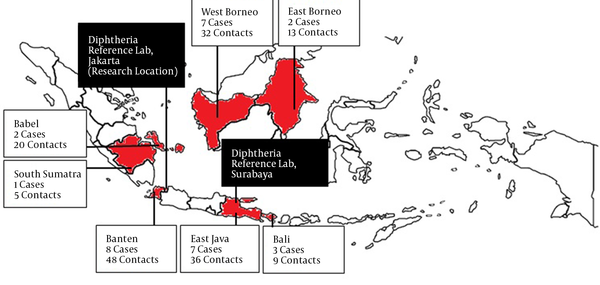
Figure 1 describes the samples based on the area of origin of sample collection and sample criteria (case or close contact). The case to close contact sample ratio was about 1 to 5, and the samples were mainly collected from Banten, East Java, and West Borneo.
Location (province) of sample collection
4.2. Laboratory Examination and Data Analysis
Out of 178 samples, 139 cases showed suspected colonies (blackish color on CTBA medium or bright white color in blood agar + fosfomycin medium) and were selected for microscopic examination and biochemical tests. Eight samples were positive for C. diphtheriae by the biochemical test. Eight positive samples were determined as toxigenic strains by the modified Elek test. On the other hand, based on the PCR assay, 17 samples were identified as toxigenic C. diphtheriae, including the eight positive samples identified by the conventional method (Table 1).
| Results | Conventional Methods | PCR | ||
|---|---|---|---|---|
| Case | Contact | Case | Contact | |
| Toxigenic Corynebacterium diphtheriae | 4 (13.8) | 4 (2.7) | 8 (27.6) | 9 (6.0) |
| Non-toxigenic C. diphtheriae | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| C. ulcerans | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| C. pseudotuberculosis | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Non-potentially toxigenic Corynebacterium | 25 (86.2) | 145 (97.3) | 21 (72.4) | 140 (94.0) |
| Total | 29 (100) | 149 (100) | 29 (100) | 149 (100) |
Toxigenic C. diphtheriae was found in 4 (13.8%) out of 29 suspected cases by the conventional method versus 8 (27.6%) out of 29 suspected cases by the PCR assay. The bacteria were also found in 4 (2.7%) out of 149 close contacts by the conventional method and 9 (6.0%) out of 149 close contacts by the PCR assay. Statistical analysis showed that the PCR assay was about twice more sensitive than the conventional method (Table 2). There was a significant correlation between different results and > 72 hours’ specimen examination delay with a P-value of 0.04 (< 0.05) (Table 3).
Statistical Analysis to Determine Sensitivity and Specificity of Both Methods
| Variables | Diphtheria Disease | Total | |
|---|---|---|---|
| Positive Clinical Cases) | Negative (Close Contact with Negative Lab Result) | ||
| PCR assay | |||
| Positive | 8 | 0 | PPV: 8/8 = 100% |
| Negative | 21 | 140 | NPV: 140/161 = 87.0% |
| Sensitivity: 8/29 = 27.6% | Specificity: 140/140 = 100% | ||
| Conventional methods | |||
| Positive | 4 | 0 | PPV: 8/8 = 100% |
| Negative | 25 | 140 | NPV: 140/165 = 84.8% |
| Sensitivity: 4/29 = 13.6% | Specificity: 140/140 = 100% | ||
The Influence of Laboratory Examination Delay on Different Results
| Laboratory Examination Delay (h) | No. | % | Positive Results | Different Results | P-Value | |
|---|---|---|---|---|---|---|
| Conventional Methods | PCR | |||||
| < 24 | 12 | 6.7 | 1 | 1 | 0 | 0.04 |
| 25 - 48 | 52 | 29.2 | 5 | 5 | 0 | |
| 49 - 72 | 14 | 7.9 | 1 | 2 | 1 | |
| > 72 | 100 | 56.2 | 1 | 9 | 8 | |
| Total | 178 | 100 | 8 | 17 | 9 | |
5. Discussion
The samples were collected from several provinces in Indonesia (Figure 1), where diphtheria cases were more prevalent in 2016. In this study, the clinical samples as throat swabs and nasal swabs were obtained as ideal samples from suspects and their close contacts for the diphtheria laboratory examination. The swab collection from close contacts was done to identify early the cases and carriers of the disease. The early detection of cases and carriers is considerably important in disease management, especially for transmission tracking. The throat swab was the main sample for diphtheria laboratory examination because the focal infection site is generally located at the tonsils and surrounding areas. In line with this, Ott et al. reported that diphtheria-causing bacteria had a higher affinity toward throat epithelial cells than nasopharyngeal cells (12). However, other samples, especially nasopharyngeal swabs, should also be collected. The bacteria causing diphtheria were found only in nasopharyngeal swabs but not in throat swabs in some cases. In this study, positive results were obtained more from throat swabs than nasopharyngeal swabs (data not shown), similar to a previous study (13).
The conventional method for diphtheria examination started with clinical sample culture on selective media. The used semi-selective medium, blood agar + fosfomycin, was intended to increase the sensitivity of the examination because tellurite in the CTBA medium can suppress the growth of bacteria causing diphtheria (4). The Albert staining was used in this study for cell morphology examination by microscopy as the common method of diphtheria examination. Other staining methods commonly used for diphtheria examination include Neisser’s Methylene Blue staining and Gram staining (9). Diphtheria-causing bacteria show some characteristics, including metachromatic granules on one or both ends and rod-like shapes. Nevertheless, it is not easy to differentiate between the Corynebacterium genus members. Hence, a biochemical test was done using API Coryne because it was easy to use and accurate (14). The toxigenicity test was conducted using the modified Elek test as an alternative gold standard.
Similar to a previous study (10), the bacteria causing diphtheria in this study were dominated by toxigenic C. diphtheriae while C. ulcerans and C. pseudotuberculosis were not found (Table 1). This result implies that one of the factors associated with the diphtheria problem in Indonesia is immunization. The diphtheria cases caused by C. ulcerans are largely found in developed countries with high immunization coverage. A previous study reported that 75% of diphtheria cases caused by C. ulcerans happened in individuals who received complete or partial diphtheria immunization (15-18). The application of the PCR assay to identify toxigenic C. diphtheriae in the diphtheria laboratory has been acknowledged since a few years ago (19-21). The PCR assay has been developed to identify C. diphtheriae, C. ulcerans, and C. pseudotuberculosis simultaneously. The PCR technique has evolved from single to multiplex PCR and conventional to real-time PCR (6). In 1997, Nakao et al. introduced a PCR assay to be applied on clinical specimens using two primer pairs targeting the tox gene subunit A and B. The results showed that the PCR examination was good enough to detect toxigenic C. diphtheriae in clinical specimens (5). Furthermore, Williams et al. (2020) developed Corynebacterium triplex RT-PCR as a rapid and sensitive tool to screen isolates and identify probable diphtheria cases directly from clinical specimens (22).
In the field, many laboratories use PCR for the direct examination of clinical specimens for many reasons. There are several advantages to PCR, as it is faster and easier to be interpreted. In addition, PCR does not require viable bacteria; thus, it is less affected by the history of antibiotic administration and sample transportation constraints. However, PCR has several limitations. A small proportion of bacteria causing diphtheria is known as the NTTB type. In the NTTB type, the tox gene is detected by PCR and considered as toxigenic bacteria, but it is non-toxigenic phenotypically. Therefore, the toxigenicity detected by PCR should be further confirmed by the Elek test or Vero cell cytotoxicity phenotypically (7-9).
Tables 1 and 2 show the advantages of PCR, especially in terms of sensitivity. The positivity rate of diphtheria-confirmed cases was only 13.8% based on the conventional method, while it was 27.6% by the PCR assay. It was supported by statistical analysis, showing that the sensitivity of PCR was about twice the sensitivity of the conventional method. The PCR can also be used as an internal control. The culture should be repeated when the culture results are negative, but the PCR results are positive, or vice versa. PCR has a limitation to determine bacterial toxigenicity of NTTB type, even though this type is barely found in Indonesia. So far, there has been only one report of the NTTB type in Indonesia that was not related to diphtheria and its contacts (23). Besides, the NTTB type can be predicted based on DNA sequences of the tox gene (23, 24). The PCR can also be developed to predict the NTTB type based on tox gene mutations, as we used in this study (10).
Indonesia is an archipelago country; thus, the geographical factor and transport system of specimens are real obstacles, which cause specimen examinations to be delayed. As known, the delay of specimen processing, especially above 72 hours, is correlated with the differentiation of the results based on statistical analysis. Table 3 shows that only a small proportion of samples arrived in the laboratory within 24 hours, while over 50% of the samples arrived later than 72 hours of collection. In addition, two samples arrived in the reference laboratory two months after collection because of the forest fire in Borneo Island. Importantly, previous research reported that a small concentration of C. diphtheriae in Amies transport medium at room temperature could not survive for more than three days (25). On the other hand, the uncontrolled use of antibiotics without a doctor’s prescription is another problem to obtain appropriate specimens for bacterial culture in Indonesia. A previous study suggested that erythromycin use for two days causes 96% of the specimens to be not cultured (negative culture) (26). This study showed that PCR deserves to be an additional examination to the conventional method in confirming diphtheria cases in Indonesia, as well as Africa. The National Institute for Communicable Diseases, Africa, has included PCR as a confirmatory method in the diphtheria laboratory (27). It is underlined that we recommend the PCR assay as additional testing, but it is not a substitution of the conventional method as a gold standard.
5.1. Conclusions
The PCR assay is more sensitive than the conventional method to identify diphtheria-causing bacteria and may be applied as additional testing to increase the positivity rate of diphtheria-confirmed cases in Indonesia as an archipelago country, where the geographical factor and specimen transport are real obstacles.
Acknowledgements
References
-
1.
World Health Organization. Diphtheria reported cases. Geneva, Switzerland: World Health Organization; 2020. Available from: https://apps.who.int/immunization_monitoring/globalsummary/timeseries/tsincidencediphtheria.html.
-
2.
Kurniawan R, Hardhana B, Siswanti T; Yudianto. [Indonesia health profile year 2017]. Jakarta, Indonesia: Kementerian Kesehatan Republik Indonesia; 2018. Indonesian.
-
3.
Efstratiou A, Engler KH, Mazurova IK, Glushkevich T, Vuopio-Varkila J, Popovic T. Current approaches to the laboratory diagnosis of diphtheria. J Infect Dis. 2000;181 Suppl 1:S138-45. [PubMed ID: 10657205]. https://doi.org/10.1086/315552.
-
4.
Berger A, Hogardt M, Konrad R, Sing A. Detection methods for laboratory diagnosis of diphtheria. Corynebacterium diphtheriae and related toxigenic species. Dordrecht, Netherlands: Springer; 2014. p. 171-205.
-
5.
Nakao H, Popovic T. Development of a direct PCR assay for detection of the diphtheria toxin gene. J Clin Microbiol. 1997;35(7):1651-5. [PubMed ID: 9196167]. [PubMed Central ID: PMC229815]. https://doi.org/10.1128/jcm.35.7.1651-1655.1997.
-
6.
De Zoysa A, Efstratiou A, Mann G, Harrison TG, Fry NK. Development, validation and implementation of a quadruplex real-time PCR assay for identification of potentially toxigenic corynebacteria. J Med Microbiol. 2016;65(12):1521-7. [PubMed ID: 27902437]. https://doi.org/10.1099/jmm.0.000382.
-
7.
Rajamani Sekar SK, Veeraraghavan B, Anandan S, Devanga Ragupathi NK, Sangal L, Joshi S. Strengthening the laboratory diagnosis of pathogenic Corynebacterium species in the Vaccine era. Lett Appl Microbiol. 2017;65(5):354-65. [PubMed ID: 28741682]. https://doi.org/10.1111/lam.12781.
-
8.
Efstratiou A, George RC. Laboratory guidelines for the diagnosis of infections caused by Corynebacterium diphtheriae and C. ulcerans. World Health Organization. Commun Dis Public Health. 1999;2(4):250-7. [PubMed ID: 10598381].
-
9.
World Health Organization. Regional workshop to build laboratory capacity on diphtheria diagnostics Christian Medical College, Vellore, India. Geneva, Swizerland: World Health Organization; 2019.
-
10.
Muna F, Sariadji K, Rukminiati Y, Febriyana D; Sunarno; Khariri, et al. New approach for the identification of potentially toxigenic Corynebacterium sp. using a multiplex PCR assay. J Microbiol Methods. 2021;184:106198. [PubMed ID: 33713727]. https://doi.org/10.1016/j.mimet.2021.106198.
-
11.
Fitriana F, Sunarno S, Syarif AK, Karyana M, Rosana Y, Moehario LH. A new modified medium for simultaneous cystinase and elek tests of bacteria causing diphtheria. Bali Med J. 2019;8(1):334. https://doi.org/10.15562/bmj.v8i1.1231.
-
12.
Ott L, Holler M, Rheinlaender J, Schaffer TE, Hensel M, Burkovski A. Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells. BMC Microbiol. 2010;10:257. [PubMed ID: 20942914]. [PubMed Central ID: PMC2965157]. https://doi.org/10.1186/1471-2180-10-257.
-
13.
Weil LM, Williams MM, Shirin T, Lawrence M, Habib ZH, Aneke JS, et al. Investigation of a large diphtheria outbreak and cocirculation of corynebacterium pseudodiphtheriticum among forcibly displaced Myanmar Nationals, 2017-2019. J Infect Dis. 2021;224(2):318-25. [PubMed ID: 33245764]. https://doi.org/10.1093/infdis/jiaa729.
-
14.
Soto A, Zapardiel J, Soriano F. Evaluation of API Coryne system for identifying coryneform bacteria. J Clin Pathol. 1994;47(8):756-9. [PubMed ID: 7962633]. [PubMed Central ID: PMC502153]. https://doi.org/10.1136/jcp.47.8.756.
-
15.
Galazka AM, Robertson SE. Diphtheria: Changing patterns in the developing world and the industrialized world. Eur J Epidemiol. 1995;11(1):107-17. [PubMed ID: 7489768]. https://doi.org/10.1007/BF01719955.
-
16.
Saikia L, Nath R, Saikia NJ, Choudhury G, Sarkar M. A diphtheria outbreak in Assam, India. Southeast Asian J Trop Med Public Health. 2010;41(3):647-52. [PubMed ID: 20578554].
-
17.
Wagner KS, White JM, Lucenko I, Mercer D, Crowcroft NS, Neal S, et al. Diphtheria in the postepidemic period, Europe, 2000-2009. Emerg Infect Dis. 2012;18(2):217-25. [PubMed ID: 22304732]. [PubMed Central ID: PMC3310452]. https://doi.org/10.3201/eid1802.110987.
-
18.
Dias AA, Santos LS, Sabbadini PS, Santos CS, Silva Junior FC, Napoleao F, et al. Corynebacterium ulcerans diphtheria: An emerging zoonosis in Brazil and worldwide. Rev Saude Publica. 2011;45(6):1176-91. [PubMed ID: 22124745]. https://doi.org/10.1590/s0034-89102011000600021.
-
19.
Hauser D, Popoff MR, Kiredjian M, Boquet P, Bimet F. Polymerase chain reaction assay for diagnosis of potentially toxinogenic Corynebacterium diphtheriae strains: Correlation with ADP-ribosylation activity assay. J Clin Microbiol. 1993;31(10):2720-3. [PubMed ID: 8253972]. [PubMed Central ID: PMC265991]. https://doi.org/10.1128/jcm.31.10.2720-2723.1993.
-
20.
Pallen MJ, Hay AJ, Puckey LH, Efstratiou A. Polymerase chain reaction for screening clinical isolates of corynebacteria for the production of diphtheria toxin. J Clin Pathol. 1994;47(4):353-6. [PubMed ID: 8027375]. [PubMed Central ID: PMC501941]. https://doi.org/10.1136/jcp.47.4.353.
-
21.
Mikhailovich VM, Melnikov VG, Mazurova IK, Wachsmuth IK, Wenger JD, Wharton M, et al. Application of PCR for detection of toxigenic Corynebacterium diphtheriae strains isolated during the Russian diphtheria epidemic, 1990 through 1994. J Clin Microbiol. 1995;33(11):3061-3. [PubMed ID: 8576378]. [PubMed Central ID: PMC228639]. https://doi.org/10.1128/jcm.33.11.3061-3063.1995.
-
22.
Williams MM, Waller JL, Aneke JS, Weigand MR, Diaz MH, Bowden KE, et al. Detection and characterization of diphtheria toxin gene-bearing Corynebacterium species through a new real-time PCR assay. J Clin Microbiol. 2020;58(10). [PubMed ID: 32727830]. [PubMed Central ID: PMC7512153]. https://doi.org/10.1128/JCM.00639-20.
-
23.
Doyle CJ, Mazins A, Graham RM, Fang NX, Smith HV, Jennison AV. Sequence analysis of toxin gene-bearing Corynebacterium diphtheriae Strains, Australia. Emerg Infect Dis. 2017;23(1):105-7. [PubMed ID: 27983494]. [PubMed Central ID: PMC5176206]. https://doi.org/10.3201/eid2301.160584.
-
24.
Zakikhany K, Neal S, Efstratiou A. Emergence and molecular characterisation of non-toxigenic tox gene-bearing Corynebacterium diphtheriae biovar mitis in the United Kingdom, 2003-2012. Euro Surveill. 2014;19(22). [PubMed ID: 24925458]. https://doi.org/10.2807/1560-7917.es2014.19.22.20819.
-
25.
Sunarno S, Sariadji K. Teknik penyimpanan dan prospek transportasi isolat corynebacterium diphtheria menggunakan silica gel. Jurnal Biotek Medisiana Indonesia. 2017;6(2):87-95. Indonesian.
-
26.
Miller LW, Bickham S, Jones WL, Heather CD, Morris RH. Diphtheria carriers and the effect of erythromycin therapy. Antimicrob Agents Chemother. 1974;6(2):166-9. [PubMed ID: 15828187]. [PubMed Central ID: PMC444622]. https://doi.org/10.1128/AAC.6.2.166.
-
27.
Archary M, Cohen C, de Gouveia L, du Plessis M, McCarthy K, Mlisana K, et al. Diphtheria: NICD recommendations for diagnosis, management and public health response. USA: National Institute for Communicable Diseases; 2018.