Abstract
Background:
Carbapenems are the most commonly administered drugs for the treatment of multidrug-resistant Pseudomonas aeruginosa (MDR P. aeruginosa) infections. However, carbapenem-resistant P. aeruginosa is spreading rapidly and has led to a new threat to human health worldwide.Objectives:
The current study aimed to determine the prevalence of imipenem-resistant P. aeruginosa, detect metallo-β-lactamase (MBL)-producer isolates, and evaluate their clonal relationships in strains isolated from patients referring to the hospitals of Ardabil city, Iran.Methods:
The resistance rate to imipenem was evaluated using the disk diffusion method. Double-disk synergy test and PCR technique were used for phenotypic and genotypic screening of MBL-positive P. aeruginosa, respectively. Ultimately, enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) and multilocus sequence typing (MLST) methods were used for assessing clonal relatedness among the isolates.Results:
The prevalence of imipenem-resistant P. aeruginosa strains was estimated at 57.1% (48 out of 84 isolates). In addition, 45 (93.7%) out of 48 imipenem-resistant P. aeruginosa isolates were phenotypically screened as MBL-positive, among which 16 (35.5%) and three (6.6%) isolates harbored blaIMP and blaVIM-1 genes, respectively. However, blaNDM, blaSIM-2, blaSPM, and blaGIM-1 genes were not detected in this study. MBL-producing P. aeruginosa strains were divided into 42 ERIC-PCR types. Based on the results of MLST, P. aeruginosa ST235 was the only identified sequence type.Conclusions:
Our results revealed a high and alarming prevalence of imipenem-resistant and blaIMP-positive P. aeruginosa ST235 at Ardabil hospitals. Continuous monitoring is essential to control the further spread of this highly virulent and drug-resistant clone.Keywords
Pseudomonas aeruginosa Imipenem Drug Resistance Metallo-β-Lactamase Clonal Relation
1. Background
β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, are good choices for the treatment of many infectious diseases caused by clinically important Pseudomonas aeruginosa. This Gram-negative opportunistic pathogen is frequently found in hospital environments, particularly intensive care units, and patients that use devices such as ventilators and blood catheters (1-3). Today, the emergence of highly resistant P. aeruginosa, especially multidrug resistant (MDR) strains, which are associated with 13% of all hospital-acquired infections, is a global public health issue (4). The Infectious Diseases Society of America (IDSA) (2016) recommends dual empiric therapy for the treatment of MDR P. aeruginosa strains (4). However, there is also a notion that necessitates antibiotic selection based on the data of local resistance patterns (4).
Carbapenems and third-generation cephalosporins are the best available drugs against MDR bacteria (3). However, the production of β-lactamase enzymes, especially class B (zinc-dependent), in P. aeruginosa isolates can cause serious challenges for treating MDR P. aeruginosa infections (1, 3). In 2017, the World Health Organization (WHO) reported that carbapenem-resistant P. aeruginosa isolates have posed a new threat to human health (3). The total prevalence of P. aeruginosa strains resistant to imipenem and meropenem (two carbapenems) in Iran is 31.6% and 40%, respectively (5). However, there is no report on the rate of carbapenem-resistant P. aeruginosa strains and their resistance mechanisms in Ardabil, a capital city in the northwest of Iran with 578,000 inhabitants (2021). Hence, the continuous monitoring of carbapenem-resistant P. aeruginosa strains and resistance mechanisms in local strains is a necessity and urgent task. An equally important task is to find new ways of infection control and new antibiotics development (3).
Among different antibiotic resistance mechanisms in P. aeruginosa, e.g., efflux pumps, loss of porins, and production of β-lactamase enzymes (6), none has yet been investigated in clinical isolates of P. aeruginosa in Ardabil. Therefore, the current study aimed to perform phenotypic and genotypic screening for metallo-β-lactamase (MBL)-producing P. aeruginosa isolates. According to the Ambler classification scheme, four classes of β-lactamases are identified, including serine β-lactamases A, C, and D, and MBL (class B) (7). Serine β-lactamases A and D and MBL are able to hydrolyze carbapenems (carbapenemase) (8). The MBL-producing P. aeruginosa strains are associated with high morbidity and mortality rates because these organisms are resistant to all β-lactam antibiotics, except for monobactams (9). The most common MBL enzymes are Imipenemase (IMP), Verona Integrin-encoded MBL (VIM), New Delhi MBL (NDM), German Imipenemase (GIM), and Seoul Imipenemase (SIM). The MBL-coding genes are located on the plasmid and bacterial chromosomes and can disseminate among bacteria through highly mobile genetic elements (9). To impede the spread of imipenem-resistant isolates of P. aeruginosa, the investigation of the clonal relatedness of MBL-producing P. aeruginosa strains is of paramount importance.
2. Objectives
Given the importance of MBL-producing (imipenem-resistant) strains, we aimed to determine the prevalence of imipenem resistance and evaluate the genetic relatedness among MBL-positive P. aeruginosa strains using enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) and multilocus sequence typing (MLST) methods.
3. Methods
3.1. Pseudomonas aeruginosa Isolates
A total of 84 clinical isolates of P. aeruginosa were used in the present study. Bacterial isolates were collected from different specimens of patients who were referred to Ardabil hospitals from June 2019 to February 2021. The collected isolates were confirmed by standard laboratory tests, including pigment production, Gram-staining, catalase, and oxidase, as well as species-specific PCR (10). Confirmed bacteria were tested to identify imipenem resistance and MBL-positive isolates using phenotypic and genotypic methods.
3.2. Phenotypic Screening for MBL Enzymes
The Kirby-Bauer’s disk diffusion method was used to evaluate P. aeruginosa resistance to the imipenem antibiotic (10 μg, Padtan Teb, Iran). Imipenem-resistant isolates were tested by double-disk synergy test, including imipenem plus imipenem-ethylenediaminetetraacetic (IPM-EDTA) (10 μg/750 μg) (Cypress Diagnostics, Belgium) disks, for phenotypic screening of MBL-producing P. aeruginosa isolates. Briefly, a 0.5 McFarland turbidity standard (1.5 × 108 CFU/mL) of P. aeruginosa was prepared and spread on Mueller-Hinton Agar (Conda, Pronasida, Spain) as a lawn culture. Imipenem and IPM-EDTA disks were placed at a 20 mm distance from each other. A growth inhibition zone of ≥ 7 mm around the IPM-EDTA disk was considered as MBL-producing P. aeruginosa. Interpretation of disk diffusion results was done according to the Clinical and Laboratory Standards Institute (CLSI) guidelines 2018 (11). For the quality control of antimicrobial susceptibility testing disks, P. aeruginosa (ATCC 27853) standard strain was used.
3.3. Genotypic Screening for MBL Enzymes
Total genomic DNA of imipenem-resistant and MBL-producer P. aeruginosa isolates was extracted using the boiling method. The PCR details, including specific primer sequences and specific annealing temperatures are presented in Table 1, and used for the detection of MBL genes, including blaIMP, blaVIM-1, blaNDM, blaSIM-2, blaSPM, and blaGIM-1 genes. The PCR was carried out in a 25 μL volume. Each PCR mixture contained 20 μL of PCR master mix (Ampliqon, Denmark), 2 μL of primers (100 pmol, 1 µL), and 3 μL of genomic DNA. Amplified products were visualized through 1% agarose gel electrophoresis and confirmed by sequencing.
Primer Sequences Used for Amplifying Metallo-β-lactamase Genes in the Present Study
| Gene | Primer Sequence (5′ to 3′) | Annealing Temperature | Amplicon Size (Bp) | Reference |
|---|---|---|---|---|
| blaIMP | F: GAAGGCGTTTATGTTCATAC | 58°C | 587 | (12) |
| R: GTACGTTTCAAGAGTGATGC | ||||
| blaVIM-1 | F: AGTGGTGAGTATCCGACAG | 52°C | 261 | (13) |
| R: ATGAAAGTGCGTGGAGAC | ||||
| blaNDM | F:GCAGCTTGTCGGCCATGCGGGC | 59°C | 782 | (12) |
| R:GGTCGCGAAGCTGAGCACCGCAT | ||||
| blaSIM-2 | F: TACAAGGGATTCGGCATCG | 52°C | 570 | (14) |
| R: TAATGGCCTGTTCCCATGTG | ||||
| blaSPM | F: GCGTTTTGTTTGCTC | 52°C | 786 | (15) |
| R:TTGGGGATGTGAGACTAC | ||||
| blaGIM-1 | F: TCGACACACCTTGGTCTGAA | 52°C | 477 | (16) |
| R: AACTTCCAACTTTGCCATGC | ||||
| ERIC-R | R: ATGTAAGCTCCTGGGGATTCAC | 48°C | - | (17) |
3.4. Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction
Clonal relatedness between MBL-producing P. aeruginosa strains was assessed using the ERIC-PCR method. The ERIC-PCR profile of each MBL-producing P. aeruginosa isolate was determined through the oligonucleotide primers and specific annealing temperature presented in Table 1. Each PCR mixture contained master mix (20 μL), template DNA (1 μL), and ERIC1-R primer (1.5 μL) (total volume of 22.5 μL). The ERIC-PCR patterns were visualized using 2% agarose gel electrophoresis. All analyses were done by an online data analysis service (inslico.ehu.es), followed by the depiction of a phylogenetic tree.
3.5. Multilocus Sequence Typing
Molecular typing of blaIMP-positive P. aeruginosa isolates was done using the MLST technique, a DNA sequence-based typing method using seven housekeeping genes. The seven housekeeping genes were amplified by PCR method, and PCR products were analyzed using 1% agarose gel electrophoresis. Amplified products were sent for sequencing, and each P. aeruginosa isolate received a sequence type number via an online data analysis service (pubmlst.org). The primer sequences and specific annealing temperatures are indicated in Table 2.
Primer Sequences Used for Multilocus Sequence Typing of blaIMP-producing Pseudomonas aeruginosa Strains
| Protein | Gene | Primer Sequence (5′ to 3′) | Annealing Temperature | Amplicon Size (Bp) | Reference |
|---|---|---|---|---|---|
| Shikimate dehydrogenase | aroE | F: ACGATTTCCCCGGGTTC | 57.4°C | 642 | pubmlst.org |
| R: CGCGCCAGAGGAAGAAT | |||||
| Acetyl coenzyme A synthetase | acsA | F: CTGGTGTACGCCTCGCTGAC | 67°C | 836 | pubmlst.org |
| R: TAGATGCCCTGCCCCTTGAT | |||||
| GMP synthase | guaA | F: CGGCCTCGACGTGTGGATGA | 71°C | 940 | pubmlst.org |
| R: GAACGCCTGGCTGGTCTTGTGGTA | |||||
| DNA mismatch repair protein | mutL | F: AGCCTGGCAGGTGGAAAC | 66°C | 619 | pubmlst.org |
| R: CAGGGTGCCATAGAGGAAGTC | |||||
| NADH dehydrogenase I chain C, D | nuoD | F: ACCGCCACCCGTATCTG | 63°C | 1042 | pubmlst.org |
| R: TCTCGCCCATCTTGACCA | |||||
| Phosphoenolpyruvate synthase | ppsA | F: GGGTAGCAAGGCGATCAAGATG | 66.4°C | 1034 | pubmlst.org |
| R: GGTTCTCTTCTTCCGGCTCGTAG | |||||
| Anthralite synthetase component I | trpE | F: GCCGATCCCTCCGAGGAAAATG | 63°C | 919 | pubmlst.org |
| R: CCCGGCGCTTGTTGATGGTT |
3.6. Statistical Analysis
The GraphPad InStat statistical software (version 3) was used for data analysis. The Student's t-test and chi-square (χ2) were used to interpret the data. A p value of < 0.05 was considered statistically significant.
4. Results
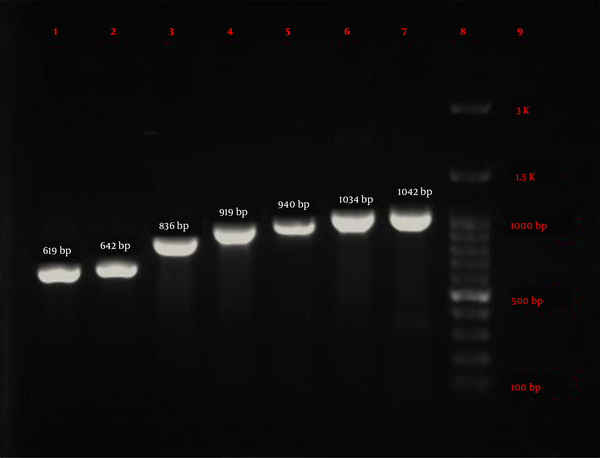
Eighty-four P. aeruginosa strains were collected from five hospitals (Alavi, Bu-Ali, Imam Reza, Imam Khomeini, and Sabalan) in Ardabil city. Based on the disk diffusion results, 48 (57.1%) out of 84 isolates were imipenem-resistant P. aeruginosa. Imipenem-resistant isolates were collected from 22 (45.9%) male (mean age 58 ± 22.6 years) and 26 (54.1%) female (mean age 55.1 ± 17.2 years) patients (P = 0.61). In addition, the distribution of imipenem-resistant P. aeruginosa strains according to the specimen type, and hospital ward were as follows: Urine 27 (56.2%), sputum 12 (25%), wound 6 (12.5%), blood 3 (6.25%), Intensive Care Unit (ICU) 13 (27%), neurology ward 13 (27%), emergency ward 7 (14.6%), internal ward 12 (25%), and pediatrics ward 3 (6.25%). Based on the double-disk synergy test, 45 (93.7%) out of 48 imipenem-resistant P. aeruginosa isolates were phenotypically screened as MBL-producing strains (Figure 1A). Additionally, among MBL-producing strains, 16 (35.5%) and three (6.6%) isolates harbored blaIMP and blaVIM-1 genes, respectively (Figure 1B). Other MBL encoding genes, i.e., blaNDM, blaSIM-2, blaSPM, and blaGIM-1 genes, were not detected in this study. The ERIC sequences of 45 MBL-producing P. aeruginosa isolates were amplified using ERIC 1R primer (Table 1) and isolates with similarities higher than 80% were considered clonally related strains. As shown in Figure 2, 42 different ERIC-PCR types were identified in MBL-producing P. aeruginosa strains. Based on MLST, seven housekeeping genes were detected using PCR (Figure 3), and ST235 was the only identified clone (Table 3). As shown in Table 3, the highest prevalence of P. aeruginosa ST235 isolation was from urine specimens (50%), Alavi hospital (68.7%), and ICU ward (37.5%).
Phenotypic and genotypic screening for MBL-producer Pseudomonas aeruginosa. (A) Double disk synergy test for phenotypic detection of MBL enzymes and (B) Agarose gel electrophoresis of amplified products of MBL genes. lane 1: IMP (587 bp), lane 2: VIM (261 bp), lane 3: Ladder (100 bp), lane 4: NDM (782 bp), lane 5: SIM-2 (570 bp), lane 6: SPM (786 bp), and lane 7: GIM (477 bp).
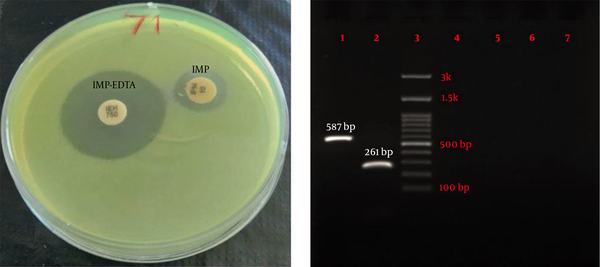
Dendrogram of ERIC-PCR patterns showing clonal relationships between MBL-producing Pseudomonas aeruginosa strains. Isolates with similarities >80% were considered clonally related strains.
Electrophoresis results of seven housekeeping genes amplified by PCR in MLST technique. lane 1: mutL (619 bp), lane 2: aroE (642 bp), lane 3: acsA (836 bp), lane 4: trpE (919 bp), lane 5: guaA (940 bp), lane 6: ppsA (1034 bp), lane 7: nuoD (1042 bp), lane 8: Ladder (100 bp), and lane 9: negative control.
Results of Multilocus Sequence Typing Technique for blaIMP-producing Pseudomonas aeruginosa Strains
| Isolate | Specimen | Hospital | Ward | Seven Housekeeping Genes | Sequence Type (ST) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| acsA | aroE | guaA | mutL | nuoD | ppsA | trpE | |||||
| 18 | Urine | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 23 | Wound | Imam Khomeini | Internal | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 24 | Urine | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 25 | Urine | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 33 | Sputum | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 34 | Sputum | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 37 | Urine | Bu-Ali | Pediatrics | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 39 | Urine | Imam Khomeini | Internal | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 43 | Urine | Alavi | ICU | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 50 | Wound | Imam Khomeini | Emergency | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 55 | Urine | Imam Reza | Internal | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 57 | Sputum | Alavi | Neurology | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 58 | Sputum | Alavi | Neurology | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 62 | Sputum | Alavi | Neurology | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 65 | Sputum | Alavi | Neurology | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
| 66 | Urine | Alavi | Neurology | 38 | 11 | 3 | 13 | 1 | 2 | 4 | 235 |
5. Discussion
Antibiotic resistance is growing rapidly worldwide. Therefore, antimicrobial resistance surveillance programs are required, especially in developing countries such as Iran, to monitor and predict the trend of drug resistance and assess the overuse and misuse of antimicrobial agents and associated consequences (18). The emergence of imipenem-resistant P. aeruginosa strains is a concerning issue because these strains show simultaneous resistance to multiple antibiotics (19). In the present study, the prevalence of imipenem-resistant P. aeruginosa strains was 57.1%, which is higher than reports from Tabriz (49%), Ahvaz (42.9%), Urmia (30.8%), Zanjan (29.2%), Guilan (23.3%), Zahedan (17.2%), and Hamadan (7.5%) and lower than in Tehran (70.4%) (5). Carbapenem-resistant P. aeruginosa prevalence in other countries has been reported as follows: Pakistan 49.5%, Philippines 31.1%, Thailand 28.7%, Japan 28.5%, Singapore 23.3%, Korea 22%, and Romania and Greece > 45%, and the Netherlands, Germany, Austria, Belgium, Denmark, Finland, French, Iceland, Luxemburg, and Malta < 20% (19).
Patients contaminated with imipenem-resistant P. aeruginosa strains experience worse treatment outcomes than those infected with imipenem-susceptible strains, causing long-term hospitalization, high costs, and substantial mortality (19). Although P. aeruginosa resistance to carbapenems is multifactorial, the production of MBL enzymes is one of the most common mechanisms (19, 20). According to the results of a meta-analysis, the total prevalence of MBL-producing P. aeruginosa in Iran was 32.4% (21). In addition, the most prevalent genes encoding MBL enzymes in Iran were blaVIM (19%) and blaIMP (11%) (21). In the current study, 53.5% (45 out of 84) of P. aeruginosa isolates were phenotypically screened as MBL-producing strains, which were divided into 42 different ERIC-PCR types based on the ERIC-PCR technique (Figure 2). On the other hand, our findings revealed that the prevalence of IMP-producing strains among imipenem-resistant P. aeruginosa clinical isolates was 35.5% in Ardabil city.
This result is higher than data from Ahvaz (0.5%, 11.7%, and 28.4%) (22-24), Zanjan (23.3% and 14.3%) (25, 26), Isfahan (31.3%) (27), Markazi (2.8%) (28), Zahedan (0.3%) (29), Kermanshah (15.1%) (30), Shiraz (3.3%) (31), and Tehran (13 %) (32). Additionally, among imipenem-resistant isolates, the rates of VIM-producing P. aeruginosa strains were 6.6% in our study, which is lower than that reported from Mashhad (11.4% and 50%), Zanjan (32%), Markazi (38%), studies from Ahvaz (8%), Kermanshah (8.3%), Tehran (33% and 13%) and Isfahan (43%, 21% and 14.6%) and higher than in Zahedan (3.7%), Shiraz (4.2%), studies from Ahvaz (0.4% and 0.8%), Kermanshah (0.9%), Tehran (0%, 0.1%, 0.5%, and 2%), and Isfahan (0.5%) (21). The blaIMP and blaVIM types are the most prevalent MBLs in the world (33). Imipenem-resistant P. aeruginosa did not contain blaNDM, blaSIM-2, blaSPM, and blaGIM-1 genes. Similar results in line with our study were reported from other cities of Iran such as Tehran, Ahvaz, Isfahan, Markazi, Zahedan, and Shiraz (21).
As known, MLST is a powerful molecular typing method for epidemiological studies of P. aeruginosa infections. According to MLST typing, P. aeruginosa sequence types (ST), ST111, ST175, ST235, ST244, and ST395, have a global distribution and are associated with outbreaks (34, 35). Among these sequence types, P. aeruginosa ST235 is the most prevalent one and is associated with highly virulent infections and multidrug resistance to carbapenems, aminoglycosides, and fluoroquinolones (34-36). Molecular typing of P. aeruginosa strains in the current study indicated that all IMP-producing isolates belonged to ST235 (Table 3). The isolation of P. aeruginosa ST235 in different hospitals in Ardabil city warns that this high-risk clone is disseminated in the investigated hospitals.
5.1. Conclusions
The high prevalence of imipenem-resistant P. aeruginosa strains in our region calls for combination therapy using synergistic antibiotics to achieve proper treatment of P. aeruginosa infections. On the other hand, considering that all IMP-producing isolates belonged to P. aeruginosa ST235, the emerging and spreading of this predominant clone in the investigated hospitals is an important challenge owing to the highly virulent and drug-resistant nature of this clone. Therefore, the continuous monitoring of drug resistance trends, resistance mechanisms, and virulence genes in local strains is essential.
References
-
1.
Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969-76. [PubMed ID: 19995920]. [PubMed Central ID: PMC2825993]. https://doi.org/10.1128/AAC.01009-09.
-
2.
Vaez H, Vaez V, Khademi F. Effect of mutation in efflux pump regulatory protein (mexr) of pseudomonas aeruginosa: A bioinformatic study. Med Lab J. 2017;11(6):35-41. https://doi.org/10.29252/mlj.11.6.35.
-
3.
World Health Organization. WHO publishes list of bacteria for which new antibiotics are urgently needed. WHO; [cited 24 October 2017]. Available from: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
-
4.
Morehead MS, Scarbrough C. Emergence of global antibiotic resistance. Prim Care. 2018;45(3):467-84. [PubMed ID: 30115335]. https://doi.org/10.1016/j.pop.2018.05.006.
-
5.
Vaez H, Salehi-Abargouei A, Ghalehnoo ZR, Khademi F. Multidrug resistant pseudomonas aeruginosa in Iran: A systematic review and metaanalysis. J Glob Infect Dis. 2018;10(4):212-7. [PubMed ID: 30581263]. [PubMed Central ID: PMC6276320]. https://doi.org/10.4103/jgid.jgid_113_17.
-
6.
Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:2475067. [PubMed ID: 27274985]. [PubMed Central ID: PMC4871955]. https://doi.org/10.1155/2016/2475067.
-
7.
Hall BG, Barlow M. Revised Ambler classification of {beta}-lactamases. J Antimicrob Chemother. 2005;55(6):1050-1. [PubMed ID: 15872044]. https://doi.org/10.1093/jac/dki130.
-
8.
Ismail SJ, Mahmoud SS. First detection of New Delhi metallo-beta-lactamases variants (NDM-1, NDM-2) among Pseudomonas aeruginosa isolated from Iraqi hospitals. Iran J Microbiol. 2018;10(2):98-103. [PubMed ID: 29997749]. [PubMed Central ID: PMC6039455].
-
9.
Farhan SM, Ibrahim RA, Mahran KM, Hetta HF, Abd El-Baky RM. Antimicrobial resistance pattern and molecular genetic distribution of metallo-beta-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect Drug Resist. 2019;12:2125-33. [PubMed ID: 31406468]. [PubMed Central ID: PMC6642648]. https://doi.org/10.2147/IDR.S198373.
-
10.
Bazghandi SA, Safarirad S, Arzanlou M, Peeri-Dogaheh H, Ali-Mohammadi H, Khademi F. Prevalence of multidrug-resistant pseudomonas aeruginosa strains in Ardabil. J Ardabil Univ Med Sci. 2021;20(2):280-6. https://doi.org/10.52547/jarums.20.2.280.
-
11.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing-twenty-eighth informational supplement. CLSI document m100. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2018. Available from: https://file.qums.ac.ir/repository/mmrc/CLSI2015.pdf.
-
12.
Hosseinzadeh Z, Sedigh Ebrahim-Saraie H, Sarvari J, Mardaneh J, Dehghani B, Rokni-Hosseini SMH, et al. Emerge of blaNDM-1 and blaOXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J Chin Med Assoc. 2018;81(6):536-40. [PubMed ID: 29030025]. https://doi.org/10.1016/j.jcma.2017.08.015.
-
13.
Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41(12):5407-13. [PubMed ID: 14662918]. [PubMed Central ID: PMC309014]. https://doi.org/10.1128/JCM.41.12.5407-5413.2003.
-
14.
Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119-23. [PubMed ID: 21398074]. https://doi.org/10.1016/j.diagmicrobio.2010.12.002.
-
15.
Ghamgosha M, Shahreki Zahedani S, Kafilzadeh F, Bameri Z. Metallo-beta-lactamase genes vim-1, Spm-1 and Imp-1 in pseudomonas aeruginosa isolated from Zahedan hospitals. Int J Infect. 2014;1(1). https://doi.org/10.17795/iji-19635.
-
16.
Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59(2):321-2. [PubMed ID: 17185300]. https://doi.org/10.1093/jac/dkl481.
-
17.
Khademi F, Maarofi K, Arzanlou M, Peeri-Dogaheh H, Sahebkar A. Which missense mutations associated with DNA gyrase and topoisomerase IV are involved in Pseudomonas aeruginosa clinical isolates resistance to ciprofloxacin in Ardabil? Gene Reports. 2021;24. https://doi.org/10.1016/j.genrep.2021.101211.
-
18.
Safdari R, GhaziSaeedi M, Masoumi-Asl H, Rezaei-Hachesu P, Mirnia K, Samad-Soltani T. A national framework for an antimicrobial resistance surveillance system within Iranian healthcare facilities: Towards a global surveillance system. J Glob Antimicrob Resist. 2017;10:59-69. [PubMed ID: 28689924]. https://doi.org/10.1016/j.jgar.2017.03.016.
-
19.
Vaez H, Salehi-Abargouei A, Khademi F. Systematic review and meta-analysis of imipenem-resistant Pseudomonas aeruginosa prevalence in Iran. Germs. 2017;7(2):86-97. [PubMed ID: 28626739]. [PubMed Central ID: PMC5466827]. https://doi.org/10.18683/germs.2017.1113.
-
20.
Rodriguez-Martinez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(11):4783-8. [PubMed ID: 19738025]. [PubMed Central ID: PMC2772299]. https://doi.org/10.1128/AAC.00574-09.
-
21.
Vaez H, Khademi F, Salehi-Abargouei A, Sahebkar A. Metallo-beta-Lactamase-producing Pseudomonas aeruginosa in Iran: a systematic review and meta-analysis. Infez Med. 2018;26(3):216-25. [PubMed ID: 30246764].
-
22.
Khosravi AD, Mihani F. Detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60(1):125-8. [PubMed ID: 17900848]. https://doi.org/10.1016/j.diagmicrobio.2007.08.003.
-
23.
Farajzadeh Sheikh A, Rostami S, Jolodar A, Tabatabaiefar MA, Khorvash F, Saki A, et al. Detection of metallo-beta lactamases among carbapenem-resistant pseudomonas aeruginosa. Jundishapur J Microbiol. 2014;7(11). e12289. [PubMed ID: 25774271]. [PubMed Central ID: PMC4332233]. https://doi.org/10.5812/jjm.12289.
-
24.
Moosavian M, Rahimzadeh M. Molecular detection of metallo-beta-lactamase genes, bla IMP-1, bla VIM-2 and bla SPM-1 in imipenem resistant Pseudomonas aeruginosa isolated from clinical specimens in teaching hospitals of Ahvaz, Iran. Iran J Microbiol. 2015;7(1):2-6. [PubMed ID: 26644866]. [PubMed Central ID: PMC4670463].
-
25.
Hemmati F, Soroori Zanjani R, Haghi F, Zeighami H. Determination of antibiotic resistance profile and frequency of metallo-beta- lactamases in pseudomonas aeruginosa isolates. J Adv Med Biomed Res. 2014;22(93):77-85.
-
26.
Doosti M, Ramazani A, Garshasbi M. Identification and characterization of metallo-beta-lactamases producing Pseudomonas aeruginosa clinical isolates in University Hospital from Zanjan Province, Iran. Iran Biomed J. 2013;17(3):129-33. [PubMed ID: 23748890]. [PubMed Central ID: PMC3770254]. https://doi.org/10.6091/ibj.1107.2013.
-
27.
Khorvash F, Yazdani M, Shabani S, Alizadeh H, Soudi A, Shoaei P, et al. Detection of Different Types of Metallo-²-Lactamases among Pseudomonas aeruginosa Isolates Obtained from Intensive Care Unit Patients. J Med Microbiol Infect Dis. 2014;2(2):84-90.
-
28.
Abdorrahim S. Molecular detection of metallo-β-lactamase genes blaVIM-1, blaVIM-2, blaIMP-1, blaIMP-2 and blaSPM-1 in Pseudomonas aeruginosa isolated from hospitalized patients in Markazi province by Duplex-PCR. African J Microbiol Res. 2012;6(12). https://doi.org/10.5897/ajmr11.1586.
-
29.
Ghamgosha M, Shahrekizahedani S, Kafilzadeh F, Bameri Z, Taheri RA, Farnoosh G. Metallo-beta-Lactamase VIM-1, SPM-1, and IMP-1 Genes Among Clinical Pseudomonas aeruginosa Species Isolated in Zahedan, Iran. Jundishapur J Microbiol. 2015;8(4). e17489. [PubMed ID: 26034547]. [PubMed Central ID: PMC4449845]. https://doi.org/10.5812/jjm.8(4)2015.17489.
-
30.
Abiri R, Mohammadi P, Shavani N, Rezaei M. Detection and genetic characterization of metallo-β-lactamase IMP-1 and VIM-2 in pseudomonas aeruginosa strains from different hospitals in Kermanshah, Iran. Jundishapur J Microbiol. 2015;8(9). https://doi.org/10.5812/jjm.22582.
-
31.
Sarhangi M, Motamedifar M, Sarvari J. Dissemination of Pseudomonas aeruginosa Producing blaIMP1, blaVIM2, blaSIM1, blaSPM1 in Shiraz, Iran. Jundishapur J Microbiol. 2013;6(7). https://doi.org/10.5812/jjm.6920.
-
32.
Sadredinamin M, Hashemi A, Goudarzi H, Tarashi S, Yousefi Nojookambari N, Taki E. Detection of blaIMP, blaVIM and OprD genes among pseudomonas aeruginosa isolated from burn patients. J Mazandaran Univ Med Sci. 2016;26(138):181-6.
-
33.
Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-58. [PubMed ID: 17630334]. [PubMed Central ID: PMC1932750]. https://doi.org/10.1128/CMR.00001-07.
-
34.
Miyoshi-Akiyama T, Tada T, Ohmagari N, Viet Hung N, Tharavichitkul P, Pokhrel BM, et al. Emergence and spread of epidemic multidrug-resistant pseudomonas aeruginosa. Genome Biol Evol. 2017;9(12):3238-45. [PubMed ID: 29202180]. [PubMed Central ID: PMC5726472]. https://doi.org/10.1093/gbe/evx243.
-
35.
Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect. 2018;24(3):258-66. [PubMed ID: 28648860]. https://doi.org/10.1016/j.cmi.2017.06.018.
-
36.
Lee AC, Jones AL. Multi-resistant Pseudomonas aeruginosa ST235 in cystic fibrosis. Paediatr Respir Rev. 2018;27:18-20. [PubMed ID: 29914746]. https://doi.org/10.1016/j.prrv.2018.05.009.