Abstract
Background:
The pandemic caused by the new coronavirus is overwhelming the world. Bacteria and fungi were detected in some cases, which suggested the associations between COVID-19 and bacterial and fungal infections.Objectives:
To provide suggestions and treatment opinions by analyzing laboratory data of COVID-19 patients co-infected with bacteria.Methods:
We analyzed 63 patients with COVID-19 admitted to the isolation ward of the First Affiliated Hospital of Wenzhou Medical University. COVID-19 was detected using PCR, and bacteria were identified using culture. Patients were divided into two groups, including those with and those without bacterial infections, and differences in hematologic indices between the groups were analyzed.Results:
There were 63 patients with median age of 55.82 years. The average hospital stay was 22.56 days. Seven patients (11.11%) had coincident bacterial infections. Detection rates in sputum/alveolar lavage and blood were the highest, 60.52% and 21.05%, respectively. Klebsiella pneumoniae, Acinetobacter, and Stenotrophomonas maltophilia were the most common found in 31.58%, 18.42%, and 15.79%, respectively. Interleukin 6 (IL-6) levels were elevated in 84.13% of patients, while IL-10 levels were elevated in 69.84%, blood ammonia levels were elevated in 82.05%, lactate levels were elevated in 75.41%, and LDH levels were elevated in 69.84%. There were significant differences between the groups in terms of expression levels of IgG, C4, AST, LDH, IL-6, IL-10, percentage of neutrophils, percentage of lymphocytes, and platelets.Conclusions:
For patients with COVID-19 suspected of having bacterial infections, empiric antibiotics should be given to cover K. pneumoniae, Acinetobacter, and S. maltophilia.Keywords
COVID-19 Co-infection Hematology Test Klebsiella pneumoniae Retrospective Studies
1. Background
At present, the pandemic caused by the new coronavirus is overwhelming the world. This new type of coronavirus emerged in Wuhan, China, in December 2019 and is named coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO) (1). COVID-19 is extremely contagious (2). As of January 28, 2021, the epidemic had spread worldwide, with nearly 100 million people were infected and more than 2.1 million died, and both numbers were rapidly increasing, with the epidemic center transferring from its origin in China to the United States (3). Wu et al. conducted an autopsy on the lungs of ten patients with COVID-19 from Wuhan and concluded that the pulmonary pathological changes of fatal COVID-19 are diffuse alveolar damage accompanied by massive neutrophil and monocyte infiltration (4). In some cases, bacteria and fungi can be detected, which suggested severe bacterial or fungal infections secondary to diffuse alveolar damage.
Several studies have shown associations between COVID-19 and bacterial infections. Rawson et al. identified 1,007 individuals on COVID-19 and reported 62/806 (8%) had bacterial/fungal co-infections (5). A retrospective study published by Zhou et al. in The Lancet showed that 28/191 (15%) hospitalized patients had bacterial infections of which 27/28 died (6). Klebsiella pneumoniae and Acinetobacter baumannii are the common pathogens identified in cultures of COVID-19 patients (7). Finally, Wang and his colleagues reported 69 patients undergoing sputum culture on admission. Of these, 5/69 (7%) had positive cultures, including Enterobacter cloacae (2/5, 50%), Candida albicans (2/5, 40%), and A. baumannii (1/5, 20%) (8). The incidence of severe bacterial infections in patients with COVID-19 is substantial. Drug-resistant bacteria such as Acinetobacter, K. pneumonia, Pseudomonas aeruginosa, multidrug-resistant Escherichia coli, Enterococcus, Chlamydia pneumoniae, and Mycoplasma pneumoniae can induce certain infections in COVID-19 patients (9-12). The use of antibacterial drugs has been reported, and approximately 72% of COVID-19 patients have received antibacterial treatment (5). It is expected that a few patients with COVID-19 will need empirical antimicrobial treatment because antibiotic therapy has shown positive therapeutic outcomes (13).
2. Objectives
For these reasons, it is critical to analyze vital signs and serum biochemical indicators of patients with COVID-19 combined with bacterial infections to achieve the best therapeutic outcomes on the premise of avoiding the overuse of antimicrobials.
3. Methods
3.1. Subjects
We gathered data on sex, age, length of stay, and laboratory test data from the hospital information system for 63 patients with COVID-19 hospitalized in the isolation ward of the First Affiliated Hospital of Wenzhou Medical University from February 1 to February 17. Thirty-nine patients tested positive for COVID-19 nucleic acid, 33 of which were throat swabs, and six were from feces; 23 were diagnosed on the basis of imaging or clinical symptoms, and one was positive for antibodies. The study was approved by the Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University.
3.2. Nucleic Acid and Microbial Detection
Throat swab and stool samples were collected for extracting SARS-CoV-2 RNA from patients who were amplified using polymerase chain reaction kits from Shengxiang Biotechnology Co., Ltd, Hunan, China. We recorded bacterial cultures from 63 patients with COVID-19 from various sources, including sputum/alveolar lavage, blood, wound drainage, feces, hydrothorax, and urine. All bacteria were identified using an automatic microbial analyzer (VITEK-2 Compact from bioMérieux, SA, France). Only the results of the first bacterial culture were retained for the same type of specimen from the same patient.
3.3. Hematologic Indices
We collected blood from patients with COVID-19 using conventional methods and evaluated hematologic indices. The hematologic indices included white blood cell count (WBC), platelet count (PLT), lymphocyte ratio, neutrophil ratio, prealbumin (PA), alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), glutamyl transpeptidase (GGT), lactate dehydrogenase (LDH), creatine kinase (CK), CK isoenzyme MB (CK-Mb), lactic acid (LAC), blood ammonia (AMON), immunoglobulin G (IgG), immunoglobulin A (IgA), immunoglobulin M (IgM), complement C3 (C3), complement C4 (C4), hypersensitive C-reactive protein (hs-CRP), chitinase 3-like protein 1 (CHI3L1), procalcitonin (PCT), interleukin 2 (IL-2), IL-4, IL-6, IL-10, tumor necrosis factor-alpha (TNF-α), interferon-gamma (IFN-γ), blood pH, blood oxygen content pressure (PO2), and blood oxygen saturation.
3.4. Statistical Analysis
When the data were normally distributed, we compared the means of continuous variables using independent T-test; otherwise, we use the Mann-Whitney U test. SPSS version 22.0 was used for data analysis, and P-values less than 0.05 were considered statistically significant. We used R Project 3.5.3 to generate all box diagrams.
4. Results
4.1. Epidemiological and Clinical Characteristics
A total of 63 patients, including 40 males (63.49%), were enrolled. The median age was 55.82 years, range 18 - 79 years, and the average hospital stay was 22.56 days (Table 1).
. Clinical Characteristics and Laboratory Hematology Indices of 63 COVID-19 Patients
| Item | Unit | No Secondary Infection | Secondary Infection | P-Value a |
|---|---|---|---|---|
| Length of stay | Day | 21.93 ± 7.22 | 27.57 ± 15.81 | 0.101 |
| Sex | 1.39 ± 0.49 | 1.14 ± 0.38 | 0.201 | |
| Age | Year | 54.64 ± 14.72 | 65.29 ± 19.00 | 0.086 |
| IgG | mg/mL | 13.14 ± 5.03 | 19.70 ± 8.87 | 0.008 |
| IgA | mg/mL | 2.83 ± 1.56 | 2.96 ± 0.41 | 0.843 |
| IgM | mg/mL | 1.05 ± 1.03 | 1.13 ± 0.32 | 0.848 |
| C3 | mg/mL | 1.07 ± 0.18 | 1.00 ± 0.16 | 0.398 |
| C4 | mg/mL | 0.38 ± 0.13 | 0.18 ± 0.04 | 0.004* |
| Prealbumin | mg/L | 148.08 ± 65.74 | 193.14 ± 79.84 | 0.136 |
| ALT | U/L | 41.65 ± 40.54 | 64.29 ± 44.72 | 0.174 |
| AST | U/L | 40.32 ± 25.92 | 76.57 ± 41.28 | 0.002* |
| ALP | U/L | 66.16 ± 30.51 | 76.57 ± 34.41 | 0.404 |
| GGT | U/L | 73.82 ± 81.49 | 113.71 ± 126.82 | 0.257 |
| LDH | U/L | 328.75 ± 125.18 | 527.29 ± 262.69 | 0.001* |
| Hs-CRP | mg/L | 34.16 ± 39.53 | 40.29 ± 28.59 | 0.697 |
| CHI3L1 | ng/ml | 180.31 ± 225.74 | 371.37 ± 205.86 | 0.09 |
| IL-2 | μg/L | 0.79 ± 0.29 | 0.99 ± 0.27 | 0.082 |
| IL-4 | μg/L | 0.80 ± 0.50 | 1.02 ± 0.42 | 0.263 |
| IL-6 | μg/L | 29.70 ± 37.37 | 80.28 ± 120.49 | 0.018* |
| IL-10 | μg/L | 7.61 ± 6.60 | 163.98 ± 406.88 | 0.004* |
| TNF-a | μg/L | 0.48 ± 0.76 | 0.38 ± 0.25 | 0.732 |
| IFN-γ | μg/L | 3.10 ± 6.13 | 1.89 ± 1.60 | 0.607 |
| WBC | × 109/L | 10.74 ± 25.60 | 10.05 ± 2.25 | 0.944 |
| Percentage of neutrophils | % | 0.64 ± 0.16 | 0.85 ± 0.07 | 0.001* |
| Percentage of lymphocytes | % | 0.24 ± 0.15 | 0.06 ± 0.04 | 0.003* |
| PLT | × 109/L | 271.84 ± 92.73 | 141.14 ± 57.51 | 0.001* |
| CK | U/L | 107.38 ± 217.35 | 255.14 ± 278.43 | 0.105 |
| CKMB | U/L | 12.40 ± 14.12 | 12.14 ± 6.34 | 0.962 |
| LAC | mmol/L | 2.77 ± 1.11 | 3.26 ± 0.92 | 0.272 |
| AMON | umol/L | 47.66 ± 28.58 | 60.00 ± 32.68 | 0.319 |
| PCT | μg/L | 0.11 ± 0.18 | 0.10 ± 0.05 | 0.824 |
| PH | 7.42 ± 0.04 | 7.44 ± 0.04 | 0.167 | |
| PO2 | mmHg | 115.14 ± 37.72 | 91.37 ± 16.37 | 0.106 |
| SPO | 97.50 ± 2.13 | 96.96 ± 1.29 | 0.515 |
4.2. Bacterial Cultures from Patients with COVID-19
Seven cases (11.11%) were combined with bacterial infection. The detection rates of sputum/alveolar lavage and blood were the highest, 60.52% and 21.05% respectively, followed by wound drainage, urine, and stool, all 5.26%, and finally hydrothorax, 2.63%. Klebsiella pneumoniae, Acinetobacter, and S. maltophilia were the most commonly isolated bacteria, with detection rates of 31.58%, 18.42%, and 15.79%, respectively. The detection rate for the Enterococcus faecalis was 7.89%, while Burkholderia multivorans, Ralstonia mannitolilytica, and E. cloacae complex were all 5.26%, and E. aerogenes, Staphylococcus aureus, P. aeruginosa, and xylose oxidation/denitrification colorless bacillus were all 2.63% (Table 2).
Results of Bacterial Culture in Different Parts of Patients with COVID-19 Secondary Bacterial Infection
| Sputum/ Alveolar Lavage | Blood | Wound Drainage | Urine | Stool | Hydrothorax | Positive Rate (%) | |
|---|---|---|---|---|---|---|---|
| Enterobacter aerogenes | 1 | 0 | 0 | 0 | 0 | 0 | 2.63 (1/38) |
| Stenotrophomonas maltophilia | 5 | 1 | 0 | 0 | 0 | 0 | 15.79 (7/38) |
| Enterococcus faecalis | 0 | 2 | 0 | 1 | 0 | 0 | 7.89 (3/38) |
| Klebsiella pneumoniae | 3 | 3 | 2 | 1 | 2 | 1 | 31.58 (12/38) |
| Staphylococcus aureus | 1 | 0 | 0 | 0 | 0 | 0 | 2.63 (1/38) |
| Burkholderia multivorans | 2 | 0 | 0 | 0 | 0 | 0 | 5.26 (2/38) |
| Acinetobacter | 6 | 1 | 0 | 0 | 0 | 0 | 18.42 (7/38) |
| Ralstonia mannitolilytica | 2 | 0 | 0 | 0 | 0 | 0 | 5.26 (2/38) |
| Enterobacter cloacae complex | 2 | 0 | 0 | 0 | 0 | 0 | 5.26 (2/38) |
| Pseudomonas aeruginosa | 1 | 0 | 0 | 0 | 0 | 0 | 2.63 (1/38) |
| Xylose oxidation/denitrification colorless bacillus | 0 | 1 | 0 | 0 | 0 | 0 | 2.63 (1/38) |
| Total, No. (%) | 23 (60.52) | 8 (21.05) | 2 (5.26) | 2 (5.26) | 2 (5.26) | 1 (2.63) |
4.3. Differences in Hematologic Indices Between Patients with or without Secondary Bacterial Infections
Interlukin-6 levels were elevated in 84.13% of patients, while IL-10 levels were elevated in 69.84%, blood ammonia levels were elevated in 82.05%, lactate levels were elevated in 75.41%, and LDH levels were elevated in 69.84% (Table 1). We compared all laboratory hematologic indices for the seven patients with secondary bacterial infection and 56 negative patients. Patients with secondary bacterial infection showed significantly higher levels of AST, LDH, IgG, IL-6, IL-10, and percentage of neutrophils (P < 0.05). By contrast, C4, percentage of lymphocytes and platelet counts were significantly lower (P < 0.05; Figure 1).
Box plot of hematology tests for the secondary infection group and the non-secondary infection group
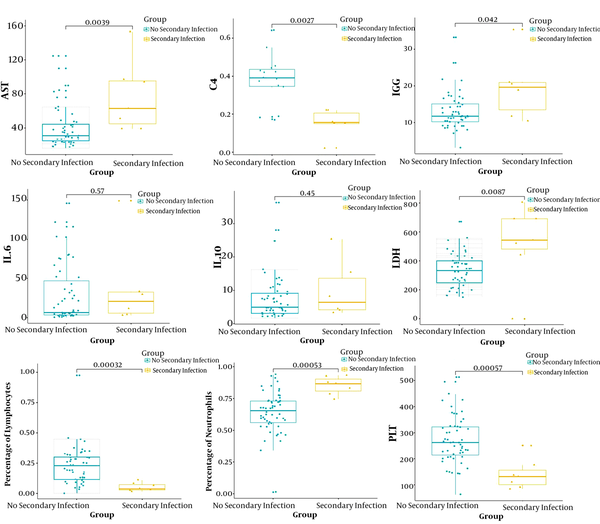
5. Discussion
This retrospective cohort study revealed that 7/63 patients had secondary bacterial infections, with an infection rate of 11.11%, slightly higher than that of a previous study (5). The possible reason was that patients admitted to this hospital were mainly severely ill. We found that the detection rate of bacteria in sputum/alveolar lavage and blood was 81.57%, and the highest percentage was in sputum/alveolar lavage alone (60.52%). This is consistent with previous reports, patients with COVID-19 who had pulmonary injuries were most likely to show bacteria in their sputum/alveolar lavage (5). We speculate that immunological or mechanical mechanisms weaken host defenses against bacteria in the respiratory tract following viral infection. Virus gives rise to the expression of new receptors for bacterial adherence and hinders ciliary clearance, resulting in colonization (14, 15).
In terms of laboratory tests, the median WBC count was elevated in both groups. The secondary bacterial infection group showed a higher neutrophil ratio, lower lymphocyte ratio, and platelet counts than the non-infected group. These findings suggest that SARS-CoV-2 may affect lymphocytes, especially T lymphocytes, as does SARS-CoV (7). When co-infected with bacteria, the virus can induce a cytokine storm, producing a series of immune responses and causing changes in peripheral WBC counts and immune cells such as lymphocytes. Patients with secondary bacterial infections had persistent and more severe lymphopenia compared with those without secondary infection, suggesting that a cellular immune deficiency state was associated with poor outcome. The neutrophil ratio in the infected group was significantly higher, which also reflected the overall inflammatory response.
Transaminases and LDH were strongly associated with COVID-19 (Table 1). In addition to bacterial infections, common complications during hospitalization were also very common (12). High levels of ALT, AST, GGT, CHI3L1, and AMON have been observed in hospitalized patients with COVID-19, especially AST, which increased more in patients with secondary bacterial infections. Studies have shown that part of the cause of elevated liver enzymes in COVID-19 patients may be the effect of lopinavir/ritonavir (16). In addition, COVID-19 mainly infects the lower respiratory tract and causes lung injury, which leads to an increase in LDH (17). Although we found that most patients with COVID-19 had significant elevations in hs-CRP, the statistical results showed that expression levels of hs-CRP did not significantly correlate with secondary bacterial infections.
SARS-COV-2 acts on bronchial epithelial cells through angiotensin-converting enzyme 2 (ACE2) receptor and induces a series of immune responses related to inflammatory cytokine storm (18, 19). Furthermore, immunoglobulin G (IgG) and cytokines produced by lymphocytes, including IL-6, IL-10, C4, are apparently elevated in severely ill patients, indicating that cytokine storm was much stronger in the severely and critically ill patients (20). There was evidence that the elevation of IL-6 levels protects the host by defending against bacteria by down-regulating the activation of the cytokine network (21, 22). Serum levels of IL-6 and IL-10 in the secondary infection group were significantly elevated, suggesting that the infected patients were in hyper-immune states during the progression of the disease, wherein there was a release of a large number of inflammatory cells and mediators.
For these reasons, we believe that IL-10, IL-6, C4, and IgG have certain advantages over traditional indicators (i.e., lymphocyte count and hs-CRP) for predicting whether patients have secondary bacterial infections. The more severe the disease, the higher the expression levels of these factors. Pneumonia is commonly associated with cardiac complications, and cardiac arrest occurs in approximately 3% of inpatients with pneumonia (23). It was also found that coronary heart disease is associated with acute cardiac events and poor outcomes in influenza and other respiratory viral infections (24, 25). This may explain why both groups of patients in the present study had higher mean levels of creatine kinase. Another finding was that the average value of lactic acid in the two groups was elevated, suggesting that patients with COVID-19 may have sepsis, septic shock, or respiratory failure.
Although viral infection can also cause sepsis syndrome, bacterial infections are generally the main cause (6). Therefore, we need to be alert to complications such as sepsis or respiratory failure caused by secondary bacterial infections in patients with COVID-19. Finally, by comparing the secondary infection and non-secondary infection groups, we found that there were no significant differences between groups in terms of gender, age, or length of stay. A possible reason is that most patients received antibiotic treatment early, which greatly shortened the length of hospital stay for patients with secondary bacterial infections. Our research has several limitations. First, because it is a retrospective study, not all laboratory tests have been performed on all patients. Therefore, the role of these factors may be underestimated in predicting secondary bacterial infections. Second, more detailed patients’ information, particularly clinical treatment, was unavailable at the time of analysis. Last but not least, the interpretation of our findings might be limited by sample size, with only seven patients with positive bacterial cultures; Nevertheless, our findings can provide suggestions regarding the serological characteristics and treatment opinions of bacterial infections secondary to SARS-CoV-2 pneumonia.
5.1. Conclusions
In summary, when patients with COVID-19 have increased levels of IgG, AST, LDH, IL-6, and IL-10, as well as elevated percentages of neutrophils and decreased levels of C4, percentage of lymphocytes, and platelets, clinicians need to be highly vigilant against bacterial infections. Cephalosporins, quinolones, and carbapenems are good initial choices for patients with COVID-19. Nevertheless, antibiotics should be de-escalated once the culture recognizes a specific organism. The “New Coronavirus Pneumonia Diagnosis and Treatment Plan (Trial Version 6)” recommends that patients with severe disease can be treated with short-term low-dose glucocorticoids as appropriate to control excessive inflammatory reactions. Simultaneously, because of medication shortages, including key antivirals, judicious use of antimicrobials will be the key to avoid bacterial resistance and ensure the maximum benefit of treatment (26, 27).
References
-
1.
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5(4):536-44. https://doi.org/10.1038/s41564-020-0695-z.
-
2.
The Lancet. Emerging understandings of 2019-nCoV. Lancet. 2020;395(10221):311. [PubMed ID: 31986259]. [PubMed Central ID: PMC7134625]. https://doi.org/10.1016/S0140-6736(20)30186-0.
-
3.
Ferrari D, Motta A, Strollo M, Banfi G, Locatelli M. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med. 2020;58(7):1095-9. https://doi.org/10.1515/cclm-2020-0398.
-
4.
Wu JH, Li X, Huang B, Su H, Li Y, Luo DJ, et al. [Pathological changes of fatal coronavirus disease 2019 (COVID-19) in the lungs: report of 10 cases by postmortem needle autopsy]. Zhonghua Bing Li Xue Za Zhi. 2020;49(6):568-75. [PubMed ID: 32486534]. https://doi.org/10.3760/cma.j.cn112151-20200405-00291.
-
5.
Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459-68. [PubMed ID: 32358954]. [PubMed Central ID: PMC7197596]. https://doi.org/10.1093/cid/ciaa530.
-
6.
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054-62. [PubMed ID: 32171076]. [PubMed Central ID: PMC7270627]. https://doi.org/10.1016/S0140-6736(20)30566-3.
-
7.
Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507-13. [PubMed ID: 32007143]. [PubMed Central ID: PMC7135076]. https://doi.org/10.1016/S0140-6736(20)30211-7.
-
8.
Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical Features of 69 Cases With Coronavirus Disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71(15):769-77. [PubMed ID: 32176772]. [PubMed Central ID: PMC7184452]. https://doi.org/10.1093/cid/ciaa272.
-
9.
Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H, et al. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9(6):496-507. [PubMed ID: 21669398]. [PubMed Central ID: PMC3125972]. https://doi.org/10.1016/j.chom.2011.05.006.
-
10.
He Y, Li W, Wang Z, Chen H, Tian L, Liu D. Nosocomial infection among patients with COVID-19: A retrospective data analysis of 918 cases from a single center in Wuhan, China. Infect Control Hosp Epidemiol. 2020;41(8):982-3. [PubMed ID: 32279676]. [PubMed Central ID: PMC7180328]. https://doi.org/10.1017/ice.2020.126.
-
11.
Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [PubMed ID: 32217556]. [PubMed Central ID: PMC7190011]. https://doi.org/10.1136/bmj.m1091.
-
12.
Wang L, He W, Yu X, Hu D, Bao M, Liu H, et al. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4-week follow-up. J Infect. 2020;80(6):639-45. [PubMed ID: 32240670]. [PubMed Central ID: PMC7118526]. https://doi.org/10.1016/j.jinf.2020.03.019.
-
13.
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506. [PubMed ID: 31986264]. [PubMed Central ID: PMC7159299]. https://doi.org/10.1016/S0140-6736(20)30183-5.
-
14.
Bragonzi A, Copreni E, de Bentzmann S, Ulrich M, Conese M. Airway epithelial cell-pathogen interactions. J Cyst Fibros. 2004;3 Suppl 2:197-201. [PubMed ID: 15463958]. https://doi.org/10.1016/j.jcf.2004.05.041.
-
15.
Wilson R, Dowling RB, Jackson AD. The biology of bacterial colonization and invasion of the respiratory mucosa. Eur Respir J. 1996;9(7):1523-30. https://doi.org/10.1183/09031936.96.09071523.
-
16.
Cai Q, Huang D, Yu H, Zhu Z, Xia Z, Su Y, et al. COVID-19: Abnormal liver function tests. J Hepatol. 2020;73(3):566-74. [PubMed ID: 32298767]. [PubMed Central ID: PMC7194951]. https://doi.org/10.1016/j.jhep.2020.04.006.
-
17.
Jurisic V, Radenkovic S, Konjevic G. The actual role of LDH as tumor marker, biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:115-24. [PubMed ID: 26530363]. https://doi.org/10.1007/978-94-017-7215-0_8.
-
18.
Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237-61. [PubMed ID: 27578435]. [PubMed Central ID: PMC5457962]. https://doi.org/10.1146/annurev-virology-110615-042301.
-
19.
Cao Y, Liu X, Xiong L, Cai K. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2: A systematic review and meta-analysis. J Med Virol. 2020;92(9):1449-59. [PubMed ID: 32242947]. [PubMed Central ID: PMC7228215]. https://doi.org/10.1002/jmv.25822.
-
20.
Wang Y, Zhu F, Wang C, Wu J, Liu J, Chen X, et al. Children hospitalized with severe COVID-19 in Wuhan. Pediatr Infect Dis J. 2020;39(7):e91-4. [PubMed ID: 32384397]. https://doi.org/10.1097/INF.0000000000002739.
-
21.
Cui W, Fan Y, Wu W, Zhang F, Wang JY, Ni AP. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37(6):857-9. [PubMed ID: 12955652]. [PubMed Central ID: PMC7110124]. https://doi.org/10.1086/378587.
-
22.
Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, et al. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol. 2002;169(8):4279-87. [PubMed ID: 12370359]. https://doi.org/10.4049/jimmunol.169.8.4279.
-
23.
Marrie TJ, Shariatzadeh MR. Community-acquired pneumonia requiring admission to an intensive care unit: a descriptive study. Medicine (Baltimore). 2007;86(2):103-11. [PubMed ID: 17435590]. https://doi.org/10.1097/MD.0b013e3180421c16.
-
24.
Corrales-Medina VF, Musher DM, Shachkina S, Chirinos JA. Acute pneumonia and the cardiovascular system. Lancet. 2013;381(9865):496-505. [PubMed ID: 23332146]. https://doi.org/10.1016/S0140-6736(12)61266-5.
-
25.
Udell JA, Zawi R, Bhatt DL, Keshtkar-Jahromi M, Gaughran F, Phrommintikul A, et al. Association between influenza vaccination and cardiovascular outcomes in high-risk patients: a meta-analysis. JAMA. 2013;310(16):1711-20. [PubMed ID: 24150467]. https://doi.org/10.1001/jama.2013.279206.
-
26.
FDA. Drug shortages. 2020, [cited 24 March 2020]. Available from: https://www.fda.gov/drugs/drug-safety-andavailability/drug-shortages.
-
27.
Rees V. EMA announces measures to manage drug shortages due to COVID-19. 2020, [cited 26 March 2020]. Available from: https://www.europeanpharmaceuticalreview.com/news/115123/ema-announces-measures-tomanage-drug-shortages-as-result-of-covid-19/.