Abstract
Background:
Leishmania is an intracellular protozoan parasite that uses complex methods for destroying the innate immune response in mammalian host macrophage cells. Many factors have been identified that play a role in the severity of the parasite’s pathogenicity. One of the factors is the GP63, which is a group of metalloproteinases that disrupts the signaling mechanism of the host cell.Objectives:
The aim of this study was to construct PX-LMGP63 vector through CRISPR-Cas9 for GP63 gene knockout in Leishmania major as a potential method for leishmanization.Methods:
A pair of gRNAs were designed based on the mRNA sequence of the GP63. Then annealing primers were cloned into the linearized vector PX-459 and transformed into the DH5ɑ competent cells. Then, PCR assay was performed with gene-specific and vector primers to confirm the colonies. In addition, the constructed plasmid was sequenced for final confirmation.Results:
The expected size band of 270 was confirmed by PCR. The plasmid sequence showed that the gRNA789 was ligated in the vector. The created structure was named PX-LMGP63 and will be transfected into the promastigote cell in the next step.Conclusions:
Owing to the prevalence of cutaneous Leishman as a public health problem in most countries and the lack of an effective vaccine for leishmaniasis, the use of the CRISPR method may make it possible to achieve an effective vaccine in the future.Keywords
1. Background
An important group of vector-borne diseases are leishmaniases that are caused by obligate protozoan parasites from the genus of Leishmania (1). The bite of infected female sandflies from the genus Lutzomyia in the New World and the genus of Phlebotomus in the Old World transmit them (2-5). Among vector-borne diseases (6, 7), leishmaniasis has the most complex and wide spectrum because it is a collection of 29 species of parasites. Leishmania includes various reservoirs and multiple vectors and is present in a wide variety of topographic foci (8-10). Cutaneous leishmaniasis, as one of the main public health problems, provides a range of diseases from self-healing infectious to chronic disfiguring disease (11).
Zinc metalloproteinase of 63 kDa is one of the main surface glycoproteins of Leishmania major, known as GP63 or leishmanolysin, which is encoded as a family with seven genes from GP63 genes 1 to 7 (12). The Leishmania GP63 gene is a potent protease that can rapidly affect host cell substrate and disrupt cell signaling. Further studies have shown its pivotal role as an important contributor to disease severity. Currently, there is no effective vaccine or medication for leishmaniasis (13). One of the main compounds against leishmaniasis is antimonial. However, toxic side effects and increasing resistance necessitate the use of alternative methods (14). Hence, recently evaluation of a vaccine against leishmaniasis was expanded.
Various vaccines such as Leishmanization, killed promastigotes, non-pathogen Leishmania, recombinant immunogenic, DNA vaccines, and various attenuated parasites have been used to protect against Leishmania (15). Virulence factors in Leishmania parasites, the most important of which are LPG and GP63, have been of interest to researchers for several years, especially in the field of vaccine and drug production. Using the GP63 gene, the parasite is able to target host body molecules, including molecules that affect signaling pathways and transcription factors and inhibit macrophage activity against the parasite. GP63 as the main receptor for macrophage leads to increased phagocytosis and its survival inside the macrophage and is necessary for attaching the parasite to the middle intestinal wall of the mosquito, and the parasite is not able to attach to the intestinal wall of the mosquito in the absence of GP63 (16-18).
CRISPR-Cas9, a recently emerged genome engineering method, has become one of the most influencing discoveries in biology (19). Clustered, regularly inter-spaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems prepare adaptive immunity against viruses and plasmids in bacteria and archaea bacteria (20). The importance of CRISPR was first found when it was recognized that some spacers corresponded to nucleotide fragments from plasmids and bacteriophage genomes. Currently, the CRISPR-Cas9 system is a beneficial gene engineering tool that is readily programmable and capable of performing DNA cleavage with the help of guide RNAs (gRNAs) and the constitutive expression of Streptococcus pyogenes-derived Cas9 nuclease (21, 22).
2. Objectives
The aim of this study was to construct a PX-LMGP63 vector through CRISPR-Cas9 for knockout GP63 gene in L. major as a potential method for leishmanization.
3. Methods
3.1. Guide RNAs Designed
Firstly, sequences of gp63 (Y00647) as target genes were achieved in FASTA format from the NCBI Gene database and Uniprot. Next, these sequences have been used as the input for the gRNA Design tool (gRNA.ctegd.uga.edu). Using the pattern of N20NGG to find the 20 bp target site, we added CACC to the 5’ end of the forward oligo and AAAC to the 5’ end of reverse oligo (Figure 1).
Illustration of design and cloning gRNA in Cas9 vector from F. Zhang Lab S. Rao and M. Zogg 05/23/2017.
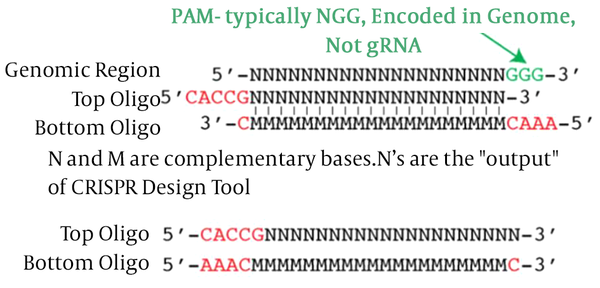
In this study for GP63 gene knockout in L. major parasite, one region of the gene was selected. A pair of gRNA was designed, and SnapGene software version 3.2.1 was used to predict the efficiency and activity of gRNAs. The designed gRNAs primer including F789: 5’-CACCGCACGAGATGGCGCACGCGCTCGG-3’ and R789: 5’-AAACCCGAGCGCGTGCGCCATCTCGTGC-3’on the GP63 gene to annealing furthermore, a pair of primers were designed to verify the accuracy of the cloned gRNA in the PX-459 vector, including ChF: 5’-GAGGGCCTATTTCCCATGATT-3’ and ChR789: 5’-CGTGCGCCATCTCGTG-3’. All oligonucleotides used in this study were synthesized by Pishgam Company, Iran.
3.2. Annealing
The components used for the annealing of oligos were 1 µM Sense Oligo, 1 µM Anti-Sense Oligo, 5 µL Annealing Buffer 10X (10 mM Tris, 1 mM EDTA, 50 mM NaCl (pH = 8.0)) and 3 µL ddH2O Total 10 µL. After making the mixture, they were annealed in a thermocycler with the program mentioned: 95°C for 5 min, then ramp down to 25°C at 1°C /min.
3.3. Vector Digestion
PX-459 is a vector that contains genes encoding SpCas9, EGFP, ampicillin resistance, and ori for the replication of the plasmid in bacterial hosts. Digestion was done with 1 µg/µL of PX-459 cloning vector with 1 µL BbsI (200U), 2 µL 10X G Buffer and X µL ddH2O up to 20 µL for overnight at 37°C to complete digestion and was heated for inactivate restriction enzyme at 65°C for 20 min. Digestion was validated on a 0.8% agarose gel electrophoresis. Linear PX459 was purified using the gel purification kit (GeneAll South Korean) then its concentration was measured by NanoDrop Analytikjena Germany.
3.4. Ligation and Transformation
To clone the annealed oligos into gRNA expression vector, ligation reaction was set up and incubated at 22°C for 3 hours. X µL digested PX-459 (20 ng), 1 µL annealing oligo duplex, 1 µL 10X T4 buffer, 0.3 µL T4 DNA ligase, X µL ddH2O (10 µL total) then were transformed to competent cell (DH5α) (23). Recombinant clones on Luria-Bertani agar plates containing ampicillin (2 µg/mL) were selected and confirmed by colony PCR (polymerase chain reactions) using GSP primers, including chF as a forward and Ch R789 as a reverse primer. All PCR assays were done in a 20 µL total volume for 35 cycles by X ng genomic DNA (100 - 200 ng for DNA and 5 - 10 ng for DNA plasmid) as a template in each reaction. The reaction mixture included 400 nM of forward and reverse primers, 10 µL Master Mix RED (AmbionTM), and the final volume was adjusted to 20 µL with ddH2O. The thermal cycler program was set as follows: the first 94°C for 5 min; followed by 35 cycles of denaturation 30 s at 94°C, annealing 30 s at 57°C and extension 30 s at 72°C and a final extension 10 min at 72°C (24). One colony was picked, and after plasmid extraction by GeneAll kit, South Korea was sequenced by Macrogen, Korea Company in one direction.
3.5. Bioinformatics
All primers were designed and checked by gRNA.ctegd.uga.edu online tools and SnapGene software (version 3.2.1), respectively. The sequence alignment was performed by nucleotide BLAST on NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequences of gp63 (Y00647) as target genes were achieved in FASTA format from Uniprot.
4. Results
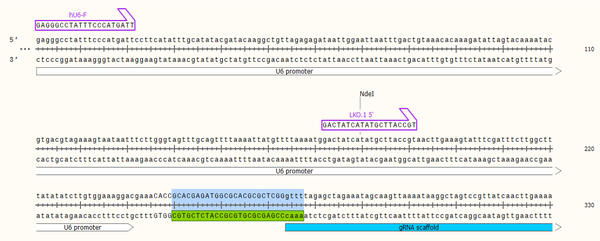
In this study, one region of the Leishmania GP63 mRNA was selected as an important point for gRNA design to knockout the gene. The gRNA was selected from Amino Acid 789 to 797. At first, using the in-silico method, cloning was done by SnapGene software (V.3.2.1) for gRNA, and it was shown the gRNA was cloned correctly (Figure 2).
In-silico cloning gRNA789 Leishmania gp63 gene by SnapGene software (V.3.2.1).
4.1. Preparation of PX-459 CRISPR Vector
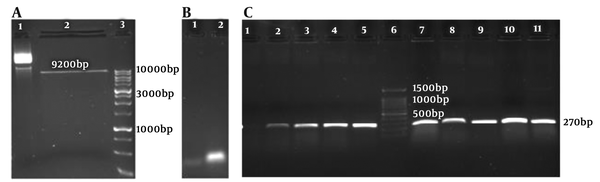
The PX-459 plasmid vector gifted by Rakhshani-Nejad was cultured; then, the plasmid was extracted. The PX-459 was digested by Bbs1 (Thermo Fisher US #ER1012) restriction enzyme (Figure 3A). The linear vector was 9200 bp. The gRNAF789 and gRNAR789 were annealed based on the mentioned protocol in material and methods (Figure 3B). The annealed gRNAs were ligated to linear PX-459 vector then transformed into Escherichia coli DH5α competent cells and cultured on LB agar plates containing 100 µL Ampicillin (50 mg/mL). After an overnight, several colonies were grown up on the plates.
A, 1: Uncut PX-459 vector; 2: linear PX-459 vector; 3: DNA marker (1 Kb); B, 1: oligonucleotide F 789; 2: gRNA789 annealed; and C, 1: No template control (NTC), 2-5 and 7-11: wells cloning PCR, 6- DNA marker (100 bp ladder).
4.2. Colony PCR
Nine colonies were picked and used as the template for colony PCR. The region containing gRNA coding DNA in the plasmid vector was amplified by chF and chR789 primers then the PCR product ran on a 1% agarose electrophoresis gel. The expected size band of 270 bp showed successful cloning of gRNA789 into PX-459 backbone plasmid vector (Figure 3C).
4.3. Plasmid Extraction and Sequencing Analysis
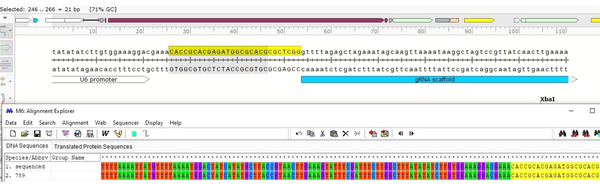
Three positive colonies were inoculated in 5 ml LB broth containing 10 µL ampicillin and incubated overnight in a shaker incubator at 37°C. Then their plasmids were extracted. After purification, plasmid colony no.: 4 was confirmed with gene-specific and vector primers, and the expected band 270 bp was observed; then, it was sequenced using the BIONEER Company (South Korea). Sequencing indicated the correct insertion of gRNA789 in the gRNA scaffold in the PX-459 vector (Figure 4). In this research, the culture medium was optimized for L. major and protocols for designing gRNA as well as annealing and cloning conditions and setting up the basic strategy for applying CRISPR-Cas9 in L. major. The designed gRNAs were cloned successfully into the pX-459 vector. The result of the present study was the production of a CRISPR structure with a gp63 gene knockout feature called PX-LMGP63, and in the next step, researchers would transfect it into a promastigote stage of the Leishmania parasite.
The sequence result indicates gRNA789 is in the frame of the PX459 vector.
5. Discussion
In the present study, a PX-LMGP63 structure was constructed that contained the gRNA789 of the GP63 gene to knockout the gene at the promastigote stage. Recently, the CRISPR-Cas9 system has been optimized for gene manipulation approaches in mammalian cells, plants, insects, and parasites (25). However, very little research has been done on adapting this system in Leishmania. In this study, we decided to optimize the CRISPR-Cas9 system in our laboratory for targeted manipulation to facilitate future gene function studies in Leishmania parasite. Vaccination with attenuated or killed parasites has not been very successful for cutaneous leishmaniasis (CL) because it has low immunogenicity and has created different immunogenicity in different areas.
With the advancement of human knowledge, the use of second-generation vaccines is on the agenda of researchers, and it is hoped that this problem can be solved in the future with the CRISPR method. The use of CRISPR targeting makes it possible to directly select the desired mutation (26). In this study, it was attempted to apply a genome-editing approach using CRISPR-Cas9 in px459 vector and utilize this system to generate a perception about its adaptability to L. major. Overall, this study facilitated the way for adapting the CRISPR-Cas9 system for gene function studies, genome editing, and creating new drugs and vaccination for future ideas. However, CRISPR-Cas9 is not a perfect tool and needs to be improved in several aspects.
Previous studies report that GC-rich sequences such as GP63 can yield more off-targets (27). This study used the powerful software available to identify sequences with minimal toxicity and tried to design gRNAs with high scores because the GP63 sequence was highly GC-rich, and designing high specific gRNAs was difficult. In 2019, Salehi Sangani et al. (15) designed the pX-leish from the CRISPR-Cas9 vector for the gp63 gene in L. major. In this study, constructed pX-leish was created with three features: (1) Promoters congenial with Leishmania parasites; (2) inserting antibiotic selection marker; (3) all-in-one vector designing, including all components required for CRISPR/Cas9. Also, in a similar study, Jesus-Santos et al. in 2020 showed that the deletion of LPG2 impaired the outcome of infection in human neutrophils, as presented by a pronounced diminution (~83%) in intracellular load compared to wild-type parasite infection (28).
The results of their study strengthen the importance of LPG and other PGs as virulence parameters in host-parasite interactions (28). The CRISPR-Cas9 system uniquely enables the simultaneous targeting of multiple genome sites (29). Genetic engineers can benefit greatly from these technologies, and together with DNA synthesis, improve their ability to decode gene function as well as manipulate them for biotechnological purposes. Compared to previous nuclease tools (ZFN and TALEN), CRISPR-Cas9 is more efficient with easier application and usage, mainly since the only variable module of CRISPR/Cas9 is the 20-base-long sgRNA which needs to be reprogrammed as the target changes (30).
In TALENs and ZFNs, sequence specificity is conferred by the DNA-binding domain of the protein, while gRNA mediates sequence specificity in the CRISPR-Cas9 system, no protein engineering is involved in this tool. As well, as CRISPR system can break methylated target sequence, but other tools cannot. Moreover, the protein domain engineering of the target DNA is a prerequisite for multiplexing using TALEN and ZFNs, which makes these techniques limited and unfit for multiplexing (31). In addition, a study was done by Zhang et al. in 2020 found that second-generation leishmaniasis using L. major LmCen strain edited by CRISPR is the first engineered tool leishmaniasis gene that lacks antibiotic-resistant markers. Although macrophage cells are the first host cells to enter the amastigote form of the leishmaniasis parasite, studies have shown that other cells of the innate immune system, such as natural killer (NK) cells, are also attacked by the leishmaniasis parasite (32, 33).
Leishmania parasites have always shown the best skill in survival, including infecting their hosts and reproduction. In fact, Leishmania is able to use different surface proteins as potential contributors to the disease [cysteine protease, gp63, lipophosphoglycan (LPG), glycosilinositolphospholipids (GIPLs)] to neutralize the host macrophage defense system and thus allowing its survival and progression within the phagolysosome environment (34). Interestingly, one of them is GP63 metalloprotease, and it is very important in the virulence of the disease. Studies have shown that Dok proteins play an important role in the survival of macrophages (34, 35). There is evidence that Dok proteins are cleaved by gp63 and positively contribute to the production of nitric oxide (NO) and tumor necrosis factor-alpha (TNF-α) in response to Leishmania infection (35). Dok family molecules, including Dok1, 2, and 3, are expressed in macrophage cells and play a negative role in the regulation of signaling in response to various growth factors, lipopolysaccharides, and cytokines (16).
A study conducted in 2014 by de Celis et al. (17) examined the role and destiny of Dok proteins following infection with L. major promastigotes in macrophages and showed that L. major is involved in altering signaling and host cell function due to its capacity. In addition, when GP63 was not expressed, Dok-1 was also partially recruited to phagosomes containing L. major promastigotes. Furthermore, TNF-α was reduced by Dok-1/Dok-2/interferon-γ-primed macrophages compared with wild-type (WT) macrophages (17). These results suggest that Dok proteins may be very important regulators in the macrophage response to Leishmania infection (17). Also, inactivation of Dok protein might be a common strategy used by several pathogens for their survival in the host. Therefore, it is suggested that further studies be performed on the deletion of L. major GP63 gene and its effects on Dok proteins in macrophages.
In a research, the L. major GP63 gene segment was amplified and then sub-cloned into pcDNA3 expression vector, and the new construct was transfected into mouse CT26 cell line (35). Furthermore, GP63 purified from L. donovani formulated in cationic DSPC liposomes induced a protective response to visceral leishmaniasis in BALB/c mice (16). In the present research, we set up protocols for synthesis gRNA, annealing, and ligation conditions for applying CRISPR/Cas9 in L. major to GP63 gene knockout. In the next steps, the CRISPR vector may be transfected into promastigotes and tested in animal models.
5.1. Conclusions
Cutaneous leishmaniasis is considered a neglected tropical disease. There is currently no effective vaccine or safe drug for this disease, so researchers’ effort to control this disease is a priority of the World Health Organization. Vaccination against leishmaniasis with L. major has been successfully performed but is no longer performed because it leads to occasional skin lesions; therefore, the use of second-generation vaccines based on the CRISPR method requires more laboratory studies and experiments. The PX-LMGP63 is capable of applying for gene targeting of gp63 in L. major. The CRISPR-Cas9 method is useful because it is competent in manipulating target genes by designing gRNAs using a simple technique for cloning the parts into the vector. The convenience and high efficiency of this technique may lead to the production of attenuated parasites as vaccines, identification of therapeutic targets, and functional genes. The present study was performed to develop this technique, which may provide promising ways ig
References
-
1.
Akhoundi M, Kuhls K, Cannet A, Votypka J, Marty P, Delaunay P, et al. A Historical Overview of the Classification, Evolution, and Dispersion of Leishmania Parasites and Sandflies. PLoS Negl Trop Dis. 2016;10(3). e0004349. [PubMed ID: 26937644]. [PubMed Central ID: PMC4777430]. https://doi.org/10.1371/journal.pntd.0004349.
-
2.
Laverde-Paz MJ, Echeverry MC, Patarroyo MA, Bello FJ. Evaluating the anti-leishmania activity of Lucilia sericata and Sarconesiopsis magellanica blowfly larval excretions/secretions in an in vitro model. Acta Trop. 2018;177:44-50. [PubMed ID: 28982577]. https://doi.org/10.1016/j.actatropica.2017.09.033.
-
3.
Karimian F, Vatandoost H, Rassi Y, Maleki-Ravasan N, Choubdar N, Koosha M, et al. wsp-based analysis of Wolbachia strains associated with Phlebotomus papatasi and P. sergenti (Diptera: Psychodidae) main cutaneous leishmaniasis vectors, introduction of a new subgroup wSerg. Pathog Glob Health. 2018;112(3):152-60. [PubMed ID: 29745300]. [PubMed Central ID: PMC6056827]. https://doi.org/10.1080/20477724.2018.1471438.
-
4.
Azizi K, Soltani A, Alipour H. Molecular detection of Leishmania isolated from cutaneous leishmaniasis patients in Jask County, Hormozgan Province, Southern Iran, 2008. Asian Pac J Trop Med. 2012;5(7):514-7. [PubMed ID: 22647811]. https://doi.org/10.1016/S1995-7645(12)60090-X.
-
5.
Azizi K, Davari B, Kalantari M, Fekri S. Gerbillid rodents fauna (Muridae: Gerbillinae) and detection of reservoir hosts (s) of zoonotic cutaneous leishmaniasis using a nested-PCR technique in Jask City in Hormozgan Province in 2008. Sci J Kurdistan Univ Med Sci. 2011;16(2).
-
6.
Akbarzadeh K, Rafinejad J, Alipour H, Biglarian A. Human myiasis in Fars Province, Iran. Southeast Asian J Trop Med Public Health. 2012;43(5):1205-11. [PubMed ID: 23431828].
-
7.
Hoosh-Deghati H, Dinparast-Djadid N, Moin-Vaziri V, Atta H, Raz AA, Seyyed-Tabaei SJ, et al. Composition of anopheles species collected from selected malarious areas of Afghanistan and Iran. J Arthropod Borne Dis. 2017;11(3):354-62. [PubMed ID: 29322052]. [PubMed Central ID: PMC5758631].
-
8.
Azizi K, Rassi Y, Javadian E, Motazedian MH, Asgari Q, Yaghoobi-Ershadi MR. First detection of Leishmania infantum in Phlebotomus (Larroussius) major (Diptera: Psychodidae) from Iran. J Med Entomol. 2008;45(4):726-31. [PubMed ID: 18714874]. https://doi.org/10.1603/0022-2585(2008)45[726:fdolii]2.0.co;2.
-
9.
Alipour H, Darabi H, Dabbaghmanesh T, Bonyani M. Entomological study of sand flies (Diptera: Psychodidae: Phlebotominae) in Asalouyeh, the heartland of an Iranian petrochemical industry. Asian Pac J Trop Biomed. 2014;4(Suppl 1):S242-5. [PubMed ID: 25183089]. [PubMed Central ID: PMC4025275]. https://doi.org/10.12980/APJTB.4.2014C678.
-
10.
Azizi K, Askari MB, Kalantari M, Moemenbellah-Fard MD. Molecular detection of Leishmania parasites and host blood meal identification in wild sand flies from a new endemic rural region, south of Iran. Pathog Glob Health. 2016;110(7-8):303-9. [PubMed ID: 27854189]. [PubMed Central ID: PMC5189871]. https://doi.org/10.1080/20477724.2016.1253530.
-
11.
David CV, Craft N. Cutaneous and mucocutaneous leishmaniasis. Dermatol Ther. 2009;22(6):491-502. [PubMed ID: 19889134]. https://doi.org/10.1111/j.1529-8019.2009.01272.x.
-
12.
Joshi PB, Sacks DL, Modi G, McMaster WR. Targeted gene deletion of Leishmania major genes encoding developmental stage-specific leishmanolysin (GP63). Mol Microbiol. 1998;27(3):519-30. [PubMed ID: 9489664]. https://doi.org/10.1046/j.1365-2958.1998.00689.x.
-
13.
Scott P, Novais FO. Cutaneous leishmaniasis: immune responses in protection and pathogenesis. Nat Rev Immunol. 2016;16(9):581-92. [PubMed ID: 27424773]. https://doi.org/10.1038/nri.2016.72.
-
14.
Ponte-Sucre A, Gamarro F, Dujardin JC, Barrett MP, Lopez-Velez R, Garcia-Hernandez R, et al. Drug resistance and treatment failure in leishmaniasis: A 21st century challenge. PLoS Negl Trop Dis. 2017;11(12). e0006052. [PubMed ID: 29240765]. [PubMed Central ID: PMC5730103]. https://doi.org/10.1371/journal.pntd.0006052.
-
15.
Salehi Sangani G, Jajarmi V, Khamesipour A, Mahmoudi M, Fata A, Mohebali M. Generation of a CRISPR/Cas9-Based Vector Specific for Gene Manipulation in Leishmania major. Iran J Parasitol. 2019;14(1):78-88. [PubMed ID: 31123471]. [PubMed Central ID: PMC6511583].
-
16.
Mahami-Oskouei M, Mohebali M, Spotin A, Alizadeh Z. A Review of effectual factors in the Pathogenesis of Leishmania Parasites. J Ardabil Univ Med Sci. 2018;18(3):279-97. https://doi.org/10.29252/jarums.18.3.279.
-
17.
Alvarez de Celis H, Gomez CP, Descoteaux A, Duplay P. Dok proteins are recruited to the phagosome and degraded in a GP63-dependent manner during Leishmania major infection. Microbes Infect. 2015;17(4):285-94. [PubMed ID: 25554486]. https://doi.org/10.1016/j.micinf.2014.12.011.
-
18.
Soares RP, Altoe ECF, Ennes-Vidal V, da Costa SM, Rangel EF, de Souza NA, et al. In Vitro Inhibition of Leishmania Attachment to Sandfly Midguts and LL-5 Cells by Divalent Metal Chelators, Anti-gp63 and Phosphoglycans. Protist. 2017;168(3):326-34. [PubMed ID: 28472733]. https://doi.org/10.1016/j.protis.2017.03.004.
-
19.
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401-7. [PubMed ID: 24861617]. [PubMed Central ID: PMC4086086]. https://doi.org/10.1093/nar/gku410.
-
20.
Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109(39):E2579-86. [PubMed ID: 22949671]. [PubMed Central ID: PMC3465414]. https://doi.org/10.1073/pnas.1208507109.
-
21.
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. [PubMed ID: 25430774]. https://doi.org/10.1126/science.1258096.
-
22.
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262-78. [PubMed ID: 24906146]. [PubMed Central ID: PMC4343198]. https://doi.org/10.1016/j.cell.2014.05.010.
-
23.
Alipour H, Raz A, Zakeri S, Djadid ND. Molecular characterization of matrix metalloproteinase-1 (MMP-1) in Lucilia sericata larvae for potential therapeutic applications. Electron J Biotechnol. 2017;29:47-56. https://doi.org/10.1016/j.ejbt.2017.06.007.
-
24.
Alipour H, Raz A, Dinparast Djadid N, Zakeri S. Expression of a new recombinant collagenase protein of Lucilia sericata in SF9 insect cell as a potential method for wound healing. Iran J Biotechnol. 2019;17(4). e2429. [PubMed ID: 32671126]. [PubMed Central ID: PMC7357693]. https://doi.org/10.30498/IJB.2019.92707.
-
25.
Nateghi Rostami M. CRISPR/Cas9 gene drive technology to control transmission of vector-borne parasitic infections. Parasite Immunol. 2020;42(9). e12762. [PubMed ID: 32497313]. https://doi.org/10.1111/pim.12762.
-
26.
Mianne J, Bourguignon C, Nguyen Van C, Fieldes M, Nasri A, Assou S, et al. Pipeline for the generation and characterization of transgenic human pluripotent stem cells using the CRISPR/Cas9 technology. Cells. 2020;9(5). [PubMed ID: 32466123]. [PubMed Central ID: PMC7290981]. https://doi.org/10.3390/cells9051312.
-
27.
Anderson EM, Haupt A, Schiel JA, Chou E, Machado HB, Strezoska Z, et al. Systematic analysis of CRISPR-Cas9 mismatch tolerance reveals low levels of off-target activity. J Biotechnol. 2015;211:56-65. [PubMed ID: 26189696]. https://doi.org/10.1016/j.jbiotec.2015.06.427.
-
28.
Jesus-Santos FH, Lobo-Silva J, Ramos PIP, Descoteaux A, Lima JB, Borges VM, et al. LPG2 Gene duplication in Leishmania infantum: A case for CRISPR-Cas9 gene editing. Front Cell Infect Microbiol. 2020;10:408. [PubMed ID: 32903718]. [PubMed Central ID: PMC7438834]. https://doi.org/10.3389/fcimb.2020.00408.
-
29.
Dong Z, Qin Q, Hu Z, Chen P, Huang L, Zhang X, et al. Construction of a one-vector multiplex CRISPR/Cas9 editing system to inhibit nucleopolyhedrovirus replication in silkworms. Virol Sin. 2019;34(4):444-53. [PubMed ID: 31218589]. [PubMed Central ID: PMC6687805]. https://doi.org/10.1007/s12250-019-00121-4.
-
30.
Xue VW, Wong SCC, Cho WCS. Genome-wide CRISPR screens for the identification of therapeutic targets for cancer treatment. Expert Opin Ther Targets. 2020;24(11):1147-58.
-
31.
Han HA, Pang JKS, Soh BS. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J Mol Med (Berl). 2020;98(5):615-32. [PubMed ID: 32198625]. [PubMed Central ID: PMC7220873]. https://doi.org/10.1007/s00109-020-01893-z.
-
32.
Lieke T, Nylen S, Eidsmo L, McMaster WR, Mohammadi AM, Khamesipour A, et al. Leishmania surface protein gp63 binds directly to human natural killer cells and inhibits proliferation. Clin Exp Immunol. 2008;153(2):221-30. [PubMed ID: 18713141]. [PubMed Central ID: PMC2492898]. https://doi.org/10.1111/j.1365-2249.2008.03687.x.
-
33.
Maasho K, Sanchez F, Schurr E, Hailu A, Akuffo H. Indications of the protective role of natural killer cells in human cutaneous leishmaniasis in an area of endemicity. Infect Immun. 1998;66(6):2698-704. [PubMed ID: 9596736]. [PubMed Central ID: PMC108258]. https://doi.org/10.1128/IAI.66.6.2698-2704.1998.
-
34.
Boulais J, Trost M, Landry CR, Dieckmann R, Levy ED, Soldati T, et al. Molecular characterization of the evolution of phagosomes. Mol Syst Biol. 2010;6:423. [PubMed ID: 20959821]. [PubMed Central ID: PMC2990642]. https://doi.org/10.1038/msb.2010.80.
-
35.
Silverman JM, Clos J, de'Oliveira CC, Shirvani O, Fang Y, Wang C, et al. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J Cell Sci. 2010;123(Pt 6):842-52. [PubMed ID: 20159964]. https://doi.org/10.1242/jcs.056465.
