Abstract
Background:
The main pathogen causing various nosocomial infections, especially in burn patients, is Staphylococcus aureus. An important virulence factor of this bacterium is leukocidin, which binds to white blood cells and leads to apoptosis and necrosis.Objectives:
In the present research, we aimed at investigating the prevalence of the panton-valentine leukocidin (PVL) and LucED genes in methicillin-susceptible and resistant isolates of S. aureus isolated from burn patients in Kermanshah, Iran.Methods:
Overall, 73 isolates of S. aureus were collected from burn wounds and identified by specific biochemical tests. After determining the susceptibility patterns of the isolates by the disc diffusion method, the frequencies of the PVL and LucED genes were assessed using specific primers and polymerase chain reactions.Results:
The rates of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible Staphylococcus aureus (MSSA) isolates were 58.9% (43 isolates) and 41.1% (30 isolates), respectively. In MRSA isolates, the highest antibiotic resistance was to penicillin (100%) and gentamicin (81.4%). Moreover, the frequencies of the LucED and PVL genes were 76.7% (56 isolates) and 27.4% (20 isolates), respectively, and the highest frequency of the LucED and PVL genes was observed in MRSA (81.4%) and MSSA (40%) isolates, respectively.Conclusions:
Increased multidrug resistance pattern among S. aureus isolates with leukocidin production has led to treatment failure and severe infections. The development of infections can be prevented by the identification of isolates carrying the leukocidin gene, and early detection of MRSA isolates is feasible by using the PVL gene.Keywords
1. Background
Staphylococcus aureus is one of the Gram-positive, facultatively anaerobic, and ubiquitous cocci that often colonizes the skin and mucous membranes (1, 2). Staphylococcus aureus is an important nosocomial opportunistic pathogen with colony stabilization and transient rates of approximately 20% and 30%, respectively (3). It is also a major cause of infection in burn patients. Among the pathogens causing infection in burn patients, S. aureus, which is a methicillin-resistant S. aureus (MRSA), has been proposed as a prominent cause of mortality (4-6). In recent years, the emergence of MRSA has been widely studied worldwide. It has been found that MRSA infections are the cause of 20,000 annual deaths in the United States. The frequency of MRSA infections in Iran was reported as 43% (7, 8). Methicillin-resistant S. aureus was formerly known as a hospital-acquired pathogen, but recently there has been an increasing trend in the reports of community-related MRSA (CA-MRSA). Some studies have reported that HA-MRSA strains are being replaced by CA-MRSA strains (9, 10).
Staphylococcus aureus has many virulence factors, including capsules, peptidoglycan, protein A, proteases, hemolysins, and leukocidin, which cause a wide range of problems, including wound infections, folliculitis, abscesses, and sepsis. This pathogen causes toxicity, endocarditis, bacteremia, osteomyelitis, and food poisoning (11, 12). Leukocidins are two-part virulence factors and pyrogenic super-antigenic toxins that can destroy host cell membranes and control immune reactions via the activation of immune cells (13). These bacteria produce seven different leukocidins, including Pantone-valentine Leukocidin (PVL), hemolysin gamma, Luk M, and Luk E/D. Moreover, PVL has been introduced as a virulence factor of S. aureus, a type of gamma toxin with two protein components called F (32 kDa) and S (38 kDa) that are controlled by the Lukf/PV and Luks/PV genes (14).
These toxins can spread among other Staphylococci due to transmission via bacteriophages. Their mechanism of action is against the outer membrane of polymorphonuclear cells, monocytes, and macrophages. Depending on the concentration of these toxins, they open calcium channels and cause apoptosis and necrosis of human leukocytes (15). The high prevalence of PVL has also been reported as a result of increasing MRSA strains (16). In addition, researchers have identified the PVL genes in CA-MRSA isolates and LukS/lukF-PV genes in methicillin-susceptible S. aureus (MSSA) (17). Staphylococcus aureus strains, which carry leukocidins, have a high level of virulence and are more common in infections such as furuncles, skin abscesses, and severe necrotic infections. In addition, S. aureus isolates containing these genes have a high potential for infection in burn patients due to their high antibiotic resistance, which, in turn, disrupts the healing process (18, 19).
2. Objectives
No comprehensive study has been recently conducted on the rate of leukocidin genes in S. aureus isolates in burn patients in Kermanshah, Iran. Thus, we aimed at determining the incidence of PVL and LucED genes in methicillin-susceptible and resistant isolates of S. aureus in burn patients who visited the Burn Unit of Imam Khomeini Hospital in Kermanshah.
3. Methods
3.1. Isolate Collection and Recognition
This descriptive cross-sectional study was performed for 11 months from December 2018 to November 2019. Sampling was performed from burn wounds of 73 patients in the Burn Unit of Imam Khomeini Hospital in Kermanshah who had not consumed any antibiotics according to patient reports and files for one week before hospitalization. The isolates were transferred to the laboratory in brain heart infusion (BHI) broth (Merck, Germany). After culture in blood agar and nutrient agar (Himdia, India), they were incubated at 37°C for 24 h. The isolates were identified by the approved biochemical tests such as Gram staining, oxidase disc, catalase, DNAase, coagulase, mannitol sugar fermentation (MSA), and novobiocin susceptibility test. For the definitive identification of S. aureus isolates, a PCR was carried out for the gene encoding deoxyribonuclease (nuc) (Table 1). After the ultimate recognition of S. aureus isolates, we kept them in a trypticase soy broth (TSB) containing 20% glycerol at a temperature of -70°C.
Primers and Temperature Cycles Used in Polymerase Chain Reaction
| Primer | Sequence (5’ - 3’) | 35 Cycles | Product Size, bp | ||
|---|---|---|---|---|---|
| Denaturation 92°C | Annealing 30 s | Extension 72°C | |||
| mecA | GTAGAAATGACTGAACGTCCGATAA | 60 s | 50°C | 60 s | 310 |
| CCAATTCCACATTGTTTCGGTCTAA | |||||
| PVL | ATCATTAGGTAAAATGTCTGCACATGATCCA | 30 s | 55°C | 45 s | 433 |
| GCATCAASTGTATTGGATAGCCAAAAGC | |||||
| luk-E/D | ATTCCATAGCATAAGCACTGC | 30 s | 55°C | 45 s | 269 |
| TGAAAAACCTTCAAAGTTGATACCAG | |||||
3.2. Antibiotic Susceptibility Testing
According to the guidelines put forth by the Clinical and Laboratory Standards Institute (CLSI), we used the Kirby-Bauer disc diffusion technique to perform anti-bacterial susceptibility testing. According to these studies (1, 20), the antibiotics included gentamicin (10 µg), amikacin (30), tobramycin (10 µg), rifampicin (5 µg), penicillin (5 µg), linezolid (30 µg), chloramphenicol (30 µg), clindamycin (2 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), and co-trimoxazole (25 µg). We used the disk diffusion method and antibiotic discs of oxacillin (1 µg) and cefoxitin (30 µg) according to the CLSI protocol for the identification of MRSA isolates. This was accomplished by adding a direct colony of S. aureus to Müller-Hinton broth and comparing it with the 0.5 McFarland standard. The suspension was cultured on a Müller-Hinton agar medium (Himdia, India), and then antibiotic discs were placed on the medium. After 24 h of incubation at 37°C, we measured the diameters of antibiotic growth inhibition zones and compared the obtained values with the standard CLSI tables. Those isolates resistant to at least three classes of antibiotics were regarded as Multidrug-resistant (MDR) isolates. Staphylococcus aureus ATCC 25923 was used for quality control in the antibiogram test.
3.3. Polymerase Chain Reaction
To examine the existence of the methicillin resistance gene (mecA), we carried out PCR on the studied isolates (Table 1) to investigate the frequency of PVL and LucED genes with the specific primers (21). Afterward, S. aureus NCTC13300 and ACTC49775 standard strains were utilized as the positive controls to detect the PVL and LukE/D genes. First, a total genomic DNA of the bacterium was extracted by the boiling method. For this purpose, several bacterial colonies were dissolved in 0.5 mL of sterile distilled water (DW) and centrifuged at 7,000 g for one minute, followed by five minutes of boiling and cooling. The supernatant solution was transferred to new Eppendorf tubes for PCR. The concentration of DNA samples was quantified by measuring optical density (OD) at 260 nm using a NanoDrop Synergy HTX (Bio Tek Instrument, Inc. Highland Park, USA) equal to 34 pmol/µL. Also, the DNA purity at 260/280 nm was 1.84.
The PCR of the genes was performed separately with a thermal cycler (Biorad, USA). The final volume for a typical 25 µL reaction was as follows: 12.5 µ of Master Mix (SinaClon Company, Iran), 3 µL of bacterial DNA, 1 µL of each primer, and 25 µL of DW. Moreover, the general formula started with an initial denaturation step at 97°C for 6 min, followed by 35 cycles according to Table 1, and finally, 10 minutes of final extensions at 72°C. The PCR products were analyzed on a 1.5% agarose gel at 85 V for 45 min. They were stained with an ethidium bromide solution and visualized under UV light in a gel documentation system (Biorad, USA).
3.4. Statistical Analysis
Fisher’s exact test and chi-square test were run to analyze the data in SPSS, version 20. A P-value of less than 0.05 was considered statistically significant.
4. Results
A total of 73 S. aureus samples were collected from burn patients [54 (74%) male patients and 19 (26%) female patients] with a mean age of 38.14 ± 16.9 years. All the isolates were from burn wound samples. It was found that the highest antibiotic resistance of the isolates was to penicillin (100%) and gentamicin (63%), whereas the lowest resistance was to linezolid (1.4%) and chloramphenicol (12.3%). The frequencies of MRSA and MSSA isolates were 58.9% (43 isolates) and 41.1% (30 isolates), respectively. In MRSA isolates, the highest antibiotic resistance was to penicillin (100%) and gentamicin (81.4%), whereas the lowest resistance was to linezolid (2.3%) and chloramphenicol (16.3%) (Table 2). The frequency of MDR isolates was determined to be 95.9% (70 isolates). Moreover, the PCR results showed that the highest frequency among the genes was related to the LucED gene with a frequency of 76.7% (56 isolates). In addition, the frequency of the PVL gene equaled 27.4% (20 isolates). Based on the results, there was a significant relationship (P < 0.05) between the existence of the LucED gene and resistance to some antibiotics, including amikacin, tobramycin, and ciprofloxacin (Table 3). The highest frequencies of the LucED and PVL genes were observed in MRSA (81.4%) and MSSA (40%) isolates, respectively (Table 4). Figures 1 and 2 depict the PCR results for the mecA, LucED, and PVL genes.
Results of Antibiotic Resistance Pattern of Methicillin-Resistant and Susceptible Staphylococcus aureus Isolatesa
| Antibiotics | MRSA (43 Isolates) | MSSA (30 Isolates) | P-Value | ||||
|---|---|---|---|---|---|---|---|
| R | S | I | R | S | I | ||
| Gentamicin | 35 (81.4) | 5 (11.6) | 3 (7) | 11 (36.7) | 19 (63.3) | 0 | 0.0001b |
| Amikacin | 23 (53.5) | 20 (46.5) | 0 | 9 (30) | 21 (70) | 0 | 0.039b |
| Tobramycin | 21 (48.8) | 22 (51.2) | 0 | 5 (16.7) | 23 (76.6) | 2 (6.7) | 0.149 |
| Rifampin | 21 (48.8) | 22 (51.2) | 0 | 6 (20) | 26 (80) | 0 | 0.011b |
| Penicillin | 43 (100) | 0 | 0 | 30 (100) | 0 | 0 | - |
| Linezolid | 1 (2.3) | 42 (97/7) | 0 | 0 | 30 (100) | 0 | 0.589 |
| Oxacillin | 19 (44.2) | 24 (55.8) | 0 | 14 (46.7) | 16 (53.3) | 0 | 0.511 |
| Ciprofloxacin | 20 (46.5) | 21 (48.8) | 2 (4.7) | 20 (66.7) | 10 (33.3) | 0 | 0.404 |
| Levofloxacin | 21 (48.8) | 20 (46.5) | 2 (4.7) | 10 (33.3) | 19 (63.3) | 1 (3.4) | 0.189 |
| Clindamycin | 21 (48.8) | 18 (41.9) | 4 (9.3) | 10 (33.3) | 19 (63.3) | 1 (3.4) | 0.64 |
| Chloramphenicol | 17 (16.3) | 36 (83.7) | 0 | 2 (6.7) | 28 (93.3) | 0 | 0.195 |
| Cotrimoxazole | 13 (3.2) | 30 (69.8) | 0 | 5 (16.7) | 25 (83.3) | 0 | 0.147 |
Association Between Frequency of Lukucidin Genes and Antibiotic Resistance
Association of Frequency of Lukucidin Genes with Methicillin-Resistant and Susceptible Staphylococcus aureus Isolatesa
| Genes | ||||
|---|---|---|---|---|
| LucED (N = 56) | PVL (N = .20) | |||
| Isolates (No.) | Positive | Negative | Positive | Negative |
| MRSA (43) | 35 (81.4) | 8 (18.6) | 8 (18.6) | 35 (81.4) |
| MSSA (30) | 21 (70) | 9 (30) | 12 (40) | 18 (60) |
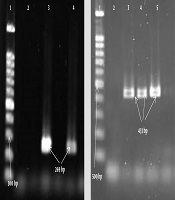
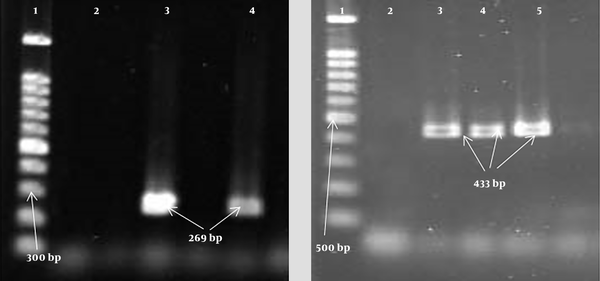
Gel electrophoresis of PCR products of leukocidin gene; LucE/D: 1, Lader (100 bp); 2, negative control; 3, positive control (269 bp); 4, positive specimen (269 bp); PVL: 1, Lader (100 bp); 2, negative control; 3, positive control (433 bp); 4, 5, positive specimen (433 bp).
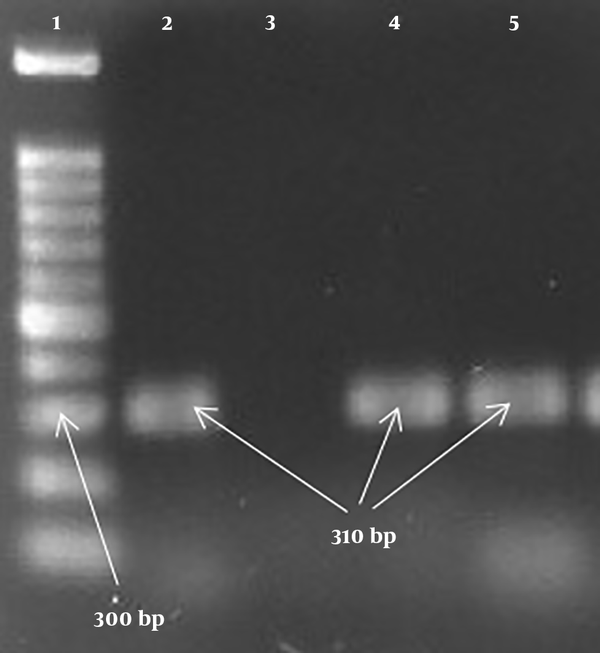
PCR products of the mecA gene: 1, Lader (100 bp); 2, positive control (310 bp); 3, negative control; 4, 5, positive specimen (310 bp).
5. Discussion
Methicillin-resistant S. aureus can be isolated from nosocomial infections. Moreover, it has long been known to cause serious infections in burn patients and escalate their mortality rates. According to the analyses conducted in this study, the frequency rates of MRSA and MSSA isolates were 58.9% and 41.1%, respectively. Concerning studies performed in Iran, the prevalence of MRSA ranged from 29% to 40% in non-burn patients and 60% to 90% in burn patients, which indicates a higher prevalence of MRSA strains in burn patients than in non-burn ones. Indeed, these results are in line with our findings (22-26).
Other studies conducted abroad have shown different results regarding the prevalence of MRSA. A study conducted by Jiang et al. on burn samples for five years revealed that out of 259 S. aureus isolates, 239 (92.28%) were MRSA, which is higher than our finding. Also, in Bangladesh, Australia, and China, the rates of MRSA were much lower than our obtained results (27-29). In MRSA isolates in this research, the highest antibiotic resistance was to penicillin (100%) and gentamicin (81.4%), and the highest sensitivity was observed to linezolid (97.7%) and chloramphenicol (83.7%). Other studies in Iran and abroad have reported high levels of resistance in MRSA isolates to different antibiotics, including penicillin, gentamicin, and ciprofloxacin (30-32). In a study carried out by Moosavian et al. (25), all MRSA isolates were sensitive to linezolid and vancomycin (25).
In this study, among 73 S. aureus isolates obtained from burn samples, the frequency rates of LucED and PVL genes were determined to be 76.7% and 27.4%, respectively. In two studies conducted in Iran that were similar to our study, the prevalence of LucED was observed to be higher than that of PVL. Khosravi et al. (22) reported that the prevalence rates of these two genes in S. aureus isolates isolated from burn samples were 66.26% and 7.2%, respectively. In addition, Havaei et al. (21) reported the frequencies of 73% and 24.8% for LucED and PVL, respectively, which are consistent with our results. Methicillin-resistant S. aureus strains carrying the PVL gene cause health problems; thus, they should be detected quickly. The first MRSA strain to carry PVL was observed in the late 1990s, and it has spread worldwide in recent years. Furthermore, PVL is a virulence factor that is transmitted to other Staphylococci through bacteriophage and contributes to the increased virulence and pathogenicity of S. aureus (33-37). In general, studies have shown that the incidence of the PVL gene in HA-MRSA is lower than the incidence of this gene in CA-MRSA. These results are completely in line with our findings, as our samples were obtained from a hospital setting (38).
In this study, the highest frequencies of the LucED and PVL genes were observed in MRSA (81.4%) and MSSA (40%) isolates, respectively. A study by Enwuru et al. (32) showed that 100% of MRSA samples carried the LucED gene, and 98% of MSSA strains had the PVL gene. In addition, a study by Khosravi et al. (22) reported the prevalence of the LucED gene to be 66.26% among MRSA strains, and the prevalence of the PVL gene in MSSA isolates was equal to 33.3%. One of the criteria for identifying nosocomial MRSA isolates is the lower prevalence of the PVL gene. In our study, the prevalence of this gene was lower than that of LucED, indicating the incidence of nosocomial infections among burn patients.
5.1. Conclusions
Despite the higher drug resistance of MRSA isolates than MSSA isolates, the frequency of the PVL gene was higher in MSSA isolates than in MRSA isolates, which indicates that burn patients in this study had acquired these infections in the hospital. The high frequency of leukocidin genes was also explained by the inherent nature of this toxin. These results can be justified by the fact that in sites such as burn wounds, bacterial death is prevented due to the active role of white blood cells, including neutrophils in wound healing, the cascading process of inflammation, and lysis of white blood cells. Considering the potential for life-threatening infections caused by the mentioned isolates and the possibility of the early detection of MSSA isolates based on the PVL gene, a wide range of nosocomial infections could be prevented by a timely diagnosis and appropriate treatment.
Acknowledgements
References
-
1.
Afsharian M, Hemmati M, Mansouri F, Azizi M, Zamanian MH, Mohseni Afshar Z, et al. Frequency of class I and II integrons in methicillin-resistant and methicillin-sensitive Staphylococcus aureus isolates in the City of Kermanshah. Arch Clin Infect Dis. 2019;14(4). https://doi.org/10.5812/archcid.86688.
-
2.
Hasanpour Dehkordi A, Khaji L, Sakhaei Shahreza MH, Mashak Z, Safarpoor Dehkordi F, Safaee Y, et al. One-year prevalence of antimicrobial susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from raw meat. Trop Biomed. 2017;34(2):396-404. [PubMed ID: 33593020].
-
3.
Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5(2):183-95. [PubMed ID: 20473345]. [PubMed Central ID: PMC2867359]. https://doi.org/10.1586/edm.10.6.
-
4.
Mir Z, Nodeh Farahani N, Abbasian S, Alinejad F, Sattarzadeh M, Pouriran R, et al. The prevalence of exotoxins, adhesion, and biofilm-related genes in Staphylococcus aureus isolates from the main burn center of Tehran, Iran. Iran J Basic Med Sci. 2019;22(11):1267-74. [PubMed ID: 32128090]. [PubMed Central ID: PMC7038426]. https://doi.org/10.22038/ijbms.2019.34908.8291.
-
5.
Amissah NA, Buultjens AH, Ablordey A, van Dam L, Opoku-Ware A, Baines SL, et al. Methicillin resistant Staphylococcus aureus transmission in a Ghanaian Burn Unit: The importance of active surveillance in resource-limited settings. Front Microbiol. 2017;8:1906. [PubMed ID: 29056927]. [PubMed Central ID: PMC5635451]. https://doi.org/10.3389/fmicb.2017.01906.
-
6.
Abdolmaleki Z, Mashak Z, Safarpoor Dehkordi F. Phenotypic and genotypic characterization of antibiotic resistance in the methicillin-resistant Staphylococcus aureus strains isolated from hospital cockroaches. Antimicrob Resist Infect Control. 2019;8:54. [PubMed ID: 30911380]. [PubMed Central ID: PMC6416839]. https://doi.org/10.1186/s13756-019-0505-7.
-
7.
Rybak M, Lomaestro B, Rotschafer JC, Moellering RJ, Craig W, Billeter M, et al. Therapeutic monitoring of vancomycin in adult patients: A consensus review of the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, and the Society of Infectious Diseases Pharmacists. Am J Health Syst Pharm. 2009;66(1):82-98. [PubMed ID: 19106348]. https://doi.org/10.2146/ajhp080434.
-
8.
Dadashi M, Nasiri MJ, Fallah F, Owlia P, Hajikhani B, Emaneini M, et al. Methicillin-resistant Staphylococcus aureus (MRSA) in Iran: A systematic review and meta-analysis. J Glob Antimicrob Resist. 2018;12:96-103. [PubMed ID: 28941791]. https://doi.org/10.1016/j.jgar.2017.09.006.
-
9.
Ahmadi E, Khojasteh M, Mortazavi SM, Khan-Mohammadi F, Kazemnia A, Beheshtipour J, et al. Prevalence of and risk factors for methicillin-resistant Staphylococcus aureus nasal carriage in the West of Iran: A population-based cross-sectional study. BMC Infect Dis. 2019;19(1):899. [PubMed ID: 31660878]. [PubMed Central ID: PMC6819401]. https://doi.org/10.1186/s12879-019-4567-1.
-
10.
Askari E, Soleymani F, Arianpoor A, Tabatabai SM, Amini A, Naderinasab M. Epidemiology of mecA-methicillin resistant Staphylococcus aureus (MRSA) in Iran: A systematic review and meta-analysis. Iran J Basic Med Sci. 2012;15(5):1010-9. [PubMed ID: 23493646]. [PubMed Central ID: PMC3586924].
-
11.
Mostafa M, Siadat SD, Shahcheraghi F, Vaziri F, Japoni-Nejad A, Vand Yousefi J, et al. Variability in gene cassette patterns of class 1 and 2 integrons associated with multi drug resistance patterns in Staphylococcus aureus clinical isolates in Tehran-Iran. BMC Microbiol. 2015;15:152. [PubMed ID: 26228695]. [PubMed Central ID: PMC4521504]. https://doi.org/10.1186/s12866-015-0488-3.
-
12.
Abdolmaleki Z, Mashak Z, Safarpoor Dehkordi F. Molecular and virulence characteristics of methicillin-resistant Staphylococcus aureus bacteria recovered from Hospital Cockroaches. Jundishapur J Microbiol. 2020;12(12). https://doi.org/10.5812/jjm.98564.
-
13.
He C, Xu S, Zhao H, Hu F, Xu X, Jin S, et al. Leukotoxin and pyrogenic toxin Superantigen gene backgrounds in bloodstream and wound Staphylococcus aureus isolates from eastern region of China. BMC Infect Dis. 2018;18(1):395. [PubMed ID: 30103694]. [PubMed Central ID: PMC6090790]. https://doi.org/10.1186/s12879-018-3297-0.
-
14.
Koop G, Vrieling M, Storisteanu DM, Lok LS, Monie T, van Wigcheren G, et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci Rep. 2017;7:40660. [PubMed ID: 28106142]. [PubMed Central ID: PMC5247767]. https://doi.org/10.1038/srep40660.
-
15.
Cupane L, Pugacova N, Berzina D, Cauce V, Gardovska D, Miklasevics E. Patients with Panton-Valentine leukocidin positive Staphylococcus aureus infections run an increased risk of longer hospitalisation. Int J Mol Epidemiol Genet. 2012;3(1):48-55. [PubMed ID: 22493751]. [PubMed Central ID: PMC3316447].
-
16.
Nikbakht M, Ahangarzadeh Rezaee M, Hasani A, Nahaei MR, Sadeghi J, Jedari Seifi S. Molecular characterization and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus isolates in Tabriz, Northwest of Iran. Arch Pediatr Infect Dis. 2017;5(1). e61620. https://doi.org/10.5812/pedinfect.61620.
-
17.
Havaei SA, Poursina F, Ahmadpour M, Havaei SR, Ruzbahani M. Detection of Panton-valentine leukocidin gene isoforms of Staphylococcus aureus isolates in Al-Zahra Hospital, Isfahan-Iran. Adv Biomed Res. 2017;6:93. [PubMed ID: 28828344]. [PubMed Central ID: PMC5549547]. https://doi.org/10.4103/2277-9175.211798.
-
18.
Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B, Coleman DC. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45(8):2554-63. [PubMed ID: 17581935]. [PubMed Central ID: PMC1951240]. https://doi.org/10.1128/JCM.00245-07.
-
19.
Brown ML, O'Hara FP, Close NM, Mera RM, Miller LA, Suaya JA, et al. Prevalence and sequence variation of panton-valentine leukocidin in methicillin-resistant and methicillin-susceptible staphylococcus aureus strains in the United States. J Clin Microbiol. 2012;50(1):86-90. [PubMed ID: 22090402]. [PubMed Central ID: PMC3256719]. https://doi.org/10.1128/JCM.05564-11.
-
20.
Rahi A, Kazemeini H, Jafariaskari S, Seif A, Hosseini S, Safarpoor Dehkordi F. Genotypic and phenotypic-based assessment of antibiotic resistance and profile of staphylococcal cassette chromosome mec in the methicillin-resistant Staphylococcus aureus recovered from raw milk. Infect Drug Resist. 2020;13:273-83. [PubMed ID: 32099419]. [PubMed Central ID: PMC6996610]. https://doi.org/10.2147/IDR.S229499.
-
21.
Havaei S, Moghadam SO, Pourmand M, Faghri J. Prevalence of genes encoding bi-component leukocidins among clinical isolates of methicillin resistant Staphylococcus aureus. Iran J Public Health. 2010;39(1):8-14. [PubMed ID: 23112984]. [PubMed Central ID: PMC3468970].
-
22.
Khosravi AD, Hoveizavi H, Farshadzadeh Z. The prevalence of genes encoding leukocidins in Staphylococcus aureus strains resistant and sensitive to methicillin isolated from burn patients in Taleghani Hospital, Ahvaz, Iran. Burns. 2012;38(2):247-51. [PubMed ID: 21924558]. https://doi.org/10.1016/j.burns.2011.08.002.
-
23.
Motamedi H, Rahmat Abadi SS, Moosavian SM, Torabi M. The association of Panton-valentine leukocidin and mecA Genes in methicillin-resistant Staphylococcus aureus isolates from patients referred to educational hospitals in Ahvaz, Iran. Jundishapur J Microbiol. 2015;8(8). e22021. [PubMed ID: 26468365]. [PubMed Central ID: PMC4601108]. https://doi.org/10.5812/jjm.22021v2.
-
24.
Dormanesh B, Siroosbakhat S, Khodaverdi Darian E, Afsharkhas L. Methicillin-resistant Staphylococcus aureus isolated from various types of hospital infections in pediatrics: Panton-valentine leukocidin, Staphylococcal chromosomal cassette mec SCCmec phenotypes and antibiotic resistance properties. Jundishapur J Microbiol. 2015;8(11). e11341. [PubMed ID: 26862375]. [PubMed Central ID: PMC4741056]. https://doi.org/10.5812/jjm.11341.
-
25.
Moosavian M, Baratian Dehkordi P, Hashemzadeh M. Characterization of SCCmec, Spa types and multidrug resistant of methicillin-resistant Staphylococcus aureus isolates in Ahvaz, Iran. Infect Drug Resist. 2020;13:1033-44. [PubMed ID: 32308445]. [PubMed Central ID: PMC7154036]. https://doi.org/10.2147/IDR.S244896.
-
26.
Vahdani P, Saifi M, Aslani MM, Asarian AA, Sharafi K. Antibiotic resistant patterns in MRSA isolates from patients admitted in ICU and infectious ward. Tanaffos. 2004;3(11):37-44.
-
27.
Islam MS, Ahmed M, Rahman SR. Incidence of methicillin resistant Staphylococcus aureus in burn patients admitted to Burn Unit, Dhaka Medical. Adv Microbiol. 2013;3(6):498-503. https://doi.org/10.4236/aim.2013.36066.
-
28.
Turnidge JD, Nimmo GR, Pearson J, Gottlieb T, Collignon PJ, Australian Group on Antimicrobial R. Epidemiology and outcomes for Staphylococcus aureus bacteraemia in Australian hospitals, 2005-06: Report from the Australian Group on Antimicrobial Resistance. Commun Dis Intell Q Rep. 2007;31(4):398-403. [PubMed ID: 18268881].
-
29.
Jiang B, Yin S, You B, Gong Y, Huang G, Yang Z, et al. Antimicrobial resistance and virulence genes profiling of methicillin-resistant Staphylococcus aureus isolates in a burn center: A 5-year study. Microb Pathog. 2018;114:176-9. [PubMed ID: 29146499]. https://doi.org/10.1016/j.micpath.2017.11.020.
-
30.
Bhatta DR, Cavaco LM, Nath G, Kumar K, Gaur A, Gokhale S, et al. Association of Panton Valentine Leukocidin (PVL) genes with methicillin resistant Staphylococcus aureus (MRSA) in Western Nepal: a matter of concern for community infections (a hospital based prospective study). BMC Infect Dis. 2016;16:199. [PubMed ID: 27179682]. [PubMed Central ID: PMC4867903]. https://doi.org/10.1186/s12879-016-1531-1.
-
31.
Zamani A, Sadeghian S, Ghaderkhani J, Alikhani MY, Najafimosleh M, Taghi Goodarzi M, et al. Detection of methicillin-resistance (mec-A) gene inStaphylococcus aureus strains by PCR and determination of antibiotic susceptibility. Ann Microbiol. 2007;57(2):273-6. https://doi.org/10.1007/bf03175219.
-
32.
Enwuru NV, Adesida SA, Enwuru CA, Ghebremedhin B, Mendie UE, Coker AO. Genetics of bi-component leukocidin and drug resistance in nasal and clinical Staphylococcus aureus in Lagos, Nigeria. Microb Pathog. 2018;115:1-7. [PubMed ID: 29246634]. https://doi.org/10.1016/j.micpath.2017.12.030.
-
33.
David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616-87. [PubMed ID: 20610826]. [PubMed Central ID: PMC2901661]. https://doi.org/10.1128/CMR.00081-09.
-
34.
Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6 Suppl 1):S3-10. discussion S62-70. [PubMed ID: 16735149]. https://doi.org/10.1016/j.amjmed.2006.03.011.
-
35.
Adler A, Givon-Lavi N, Moses AE, Block C, Dagan R. Carriage of community-associated methicillin-resistant Staphylococcus aureus in a cohort of infants in southern Israel: Risk factors and molecular features. J Clin Microbiol. 2010;48(2):531-8. [PubMed ID: 20007386]. [PubMed Central ID: PMC2815591]. https://doi.org/10.1128/JCM.02290-08.
-
36.
Sit PS, Teh CS, Idris N, Sam IC, Syed Omar SF, Sulaiman H, et al. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC Infect Dis. 2017;17(1):274. [PubMed ID: 28407796]. [PubMed Central ID: PMC5390426]. https://doi.org/10.1186/s12879-017-2384-y.
-
37.
Rezazadeh M, Yousefi Mashouf R, Sarmadyan H, Ghaznavi-Rad E. Antibiotic profile of methicillin-resistant Staphylococcus aureus with multiple-drug resistances isolated from nosocomial infections in Vali-Asr Hospital of Arak. J Arak Univ Med Sci. 2013;16(2):29-37.
-
38.
Morgan MS. Diagnosis and treatment of Panton-Valentine leukocidin (PVL)-associated staphylococcal pneumonia. Int J Antimicrob Agents. 2007;30(4):289-96. [PubMed ID: 17629464]. https://doi.org/10.1016/j.ijantimicag.2007.04.019.