Abstract
Background:
Infections caused by metallo-β-lactamases (MβLs)-producing antibiotic-resistant bacteria pose a severe threat to public health. The synergistic use of current antibiotics in combination with MβL inhibitors is a promising therapeutic mode against these antibiotic-resistant bacteria.Objectives:
The study aimed to probe the inhibition of MβLs and obtain the active component, P1, in the degradation product after imipenem was hydrolyzed by ImiS.Methods:
The hydrolysis of two carbapenems with MβL ImiS was monitored by UV-Vis in real-time, and the degradation product from the leaving group produced after imipenem was hydrolyzed (but not for faropenem) was purified by HPLC to give one component, P1.Results:
Kinetic assays revealed that P1 exhibited a broad-spectrum inhibition against VIM-2, NDM-1, ImiS, and L1, from three sub-classes of MβLs, with IC50 values of 8 - 32, 13.8 - 29.3, and 14.2 - 19.2 µM, using imipenem, cefazolin, and nitrocefin as substrates, respectively. Also, P1 showed synergistic antibacterial efficacy against drug-resistant Escherichia coli producing VIM-2, NDM-1, ImiS, and L1, in combination with antibiotics, restoring 16 to 32-fold and 32 to 128-fold efficacies of imipenem and cefazolin, respectively. Spectroscopic and Ellman's reagent analyses suggested that P1, a mercaptoethyl-form imidamide, is a mechanism-based inhibitor, while faropenem has no substrate inhibition, due to the lack of a leaving group.Conclusions:
This work reveals that the hydrolysate of imipenem, a carbapenem with a good leaving group, can be used in screening for broad-spectrum inhibitors of MβLs.Keywords
Antibiotic Resistance Metallo-β-lactamase Inhibitor Synergistic Antibacterial Efficacy
1. Background
The discovery and development of several classes of safe and efficacious antibiotics, especially β-lactams, markedly reduced mortality from bacterial infections in the past decades. However, the overuse and misuse of these antibiotics, in both medicine and agriculture, have driven the rapid evolution and dissemination of antibiotic resistance (1). β-lactam antibiotic resistance is most often a consequence of the bacterial production of β-lactamases. β-Lactamases hydrolyze the β-lactam ring of antibiotics, causing them to become ineffective in treating bacterial infections (2).
So far, more than 2,000 distinct β-lactamases have been identified, and these enzymes have been categorized into four classes, A, B, C, and D, based on their amino acid sequence, mechanism of action, and other biochemical characteristics (3, 4). Class A, C, and D enzymes are collectively called serine β-lactamases (SβLs). Class B enzymes, also called metallo-β-lactamases (MβLs), have been further sub-grouped into sub-classes of B1-B3, depending on their amino acid sequence homologies and Zn (II) content (4). Enzymes from the B1 and B3 sub-classes have broad substrate profiles and require two Zn (II) ions for maximal enzymatic activity. In contrast, the sub-class B2 enzymes exhibit maximal activity when bound to only one Zn (II) (5), and have a relatively narrow substrate profile, almost exclusively hydrolyzing carbapenems (4, 5). Despite reports suggesting that mercaptan (6), dicarboxylic acid (7), biphenyl tetrazole (8), and ebselen (9) have inhibitory activity against MβLs, there has been no inhibitor available clinically to date; therefore, the development of MβL inhibitors is urgently needed.
ImiS, a representative of B2 sub-class MβLs, from Aeromonas sobria, exhibits full activity with one Zn (II) ion bound and preferentially hydrolyzes carbapenems, including imipenem and faropenem (10), which have been called “last resort” antibiotics (11). Therefore, a large amount of effort has been made into spectroscopic, mechanistic, and inhibition studies of the enzyme (12). Spectroscopic studies show that the catalytic activity of ImiS requires a Zn (II) ion, which is bound through a cysteine, histidine, and aspartic acid residue (12), and the reaction of ImiS with imipenem leads to a product-bound species (13). Site-directed mutagenesis studies revealed that Lys224 of ImiS plays a catalytic role, while side-chain Asn233 has no role in catalysis. Inhibition studies indicated that dicarboxylic acid derivatives competitively inhibit ImiS (14).
Generally, the hydrolysate of carbapenems by MβLs is a ring-open product of β-lactam (15). Steady-state kinetic studies indicated that the hydrolysis of carbapenems, including imipenem and meropenem, by ImiS results in a significant substrate inhibition, based on Michaelis-Menten plots. Crowder et al. reported that the hydrolysates of imipenem with ImiS and L1 (a B3 sub-class MβL) inhibited both ImiS and L1 and exhibited different inhibitory activities although there were no differences in the mass spectra or the NMR spectra of the two products, suggesting that the product of biapenem hydrolysis, which occurred after C-N bond cleavage, is a bicyclic product (16).
2. Objectives
To identify whether the hydrolysate of carbapenem inhibits MβLs, in this study, two antibiotics (imipenem and faropenem) were hydrolyzed with ImiS and monitored by UV-Vis. The active component of hydrolysates was confirmed, and the inhibitory activity of the active component was evaluated with MβLs. Furthermore, the antibacterial activity of the active component against Escherichia coli with MβLs was evaluated in combination with antibiotics. In addition, the cytotoxicity of the active component against L-929 mouse fibroblastic cells was tested.
3. Methods
3.1. General Information
General chemicals were purchased from TCI (Tokyo Chemical Industry, Tokyo, Japan) and used without further purification. All antibiotics used were purchased from Sigma-Aldrich (St. Louis, MO, USA). The real-time monitoring of imipenem and faropenem hydrolysis and inhibition evaluation of MβLs were performed using an Agilent-8453 UV-visible spectrometer (Santa Clara, CA, USA). The 1H NMR and 1H-13C COSY NMR spectra were recorded on a Bruker DRX 600 MHz spectrometer (Bruker Daltonics Inc., Billerica, MA, USA).
3.2. Preparation of Escherichia coli with ImiS
The plasmid pET26b (+) with the ImiS gene was transformed into E. coli BL21. The kanamycin resistance gene existed in the plasmid. The monoclone of E. coli with ImiS was inoculated into 5 mL of Luria-Bertani media in the presence of 25 µg/mL of kanamycin and grown with shaking (150 rpm) at 37°C until the cells reached OD600 = 0.5 - 0.6. At that time, 100 Μm of IPTG was added for MβL induction, and cells were grown for 2 hours at 37°C with shaking (150 rpm). Cell cultures were centrifuged at 5,500 g for 10 min at 4°C, the supernatant was discarded, and the cell pellets were washed thoroughly by re-suspending them in 1 mL of buffer (50 mM Tris at pH = 7.0). They were then pelleted again by centrifugation (5,500 g for 10 min at 4°C). This process was repeated three times, and finally, the cells were re-suspended in buffer with an OD600 of 0.10 for UV-Vis studies (17, 18).
3.3. UV-Vis Monitoring of Antibiotics Hydrolysis
Either 10 µL of imipenem (12 mM) or 10 µL of faropenem (8 mM) was titrated into the sample cell filled with 990 µL of ImiS enzyme (0.2 µM) or E. coli with ImiS with OD600 of 0.10. Then, the variation of the UV spectrum in the reaction process was monitored at room temperature. This measurement continued until the UV absorption held steady on the Agilent UV8453 UV-Vis spectrometer (17, 18).
3.4. Determination of Minimum Inhibitory Concentration
The minimum inhibitory concentration (MIC) values of the antibiotics alone and in the presence of the enzyme inhibitor against antibiotic-resistant bacteria were determined using the broth micro-dilution method (19). Single colonies of E. coli-DH10B containing plasmids pBCSK(+)-VIM-2, pBCSK(+)-NDM-1, pBCSK(+)-ImiS, or pBCSK(+)-L1, in lysogeny broth agar plates were transferred into 5 mL of Mueller-Hinton liquid medium. Strains, grown in Mueller-Hinton medium to OD600 = 0.45, were used as inocula after 84-fold dilution to 1 × 105 CFU/mL in Mueller-Hinton medium. Cefazolin was dissolved in Mueller-Hinton medium to prepare 4096, 2048, 1024, 512, 256, 128, 64, 32, 16, and 8 µg/mL stock solutions, and imipenem was dissolved in Mueller-Hinton medium to prepare 256, 128, 64, 32, 16, 8, 4, 2, 1, and 0.5 µg/mL stock solutions. As an inhibitor, P1 was dissolved in Mueller-Hinton media to prepare a 160 µg/mL stock solution. The prepared solutions, with different antibiotic concentrations (50 µL), were diluted to 100 µL with a 50 µL inhibitor solution. Then, 100 µL inoculum was added sequentially into the prepared solutions to yield a quarter of the initial concentrations. The mixtures were incubated at 37°C for 16 h. The MIC results were taken as the lowest concentration that completely inhibited visible growth. Each measurement was performed in duplicate, and the average value was recorded.
3.5. Cytotoxicity Evaluation
A cytotoxicity assay was performed to evaluate the toxicity of P1 in mouse fibroblast cells (L-929). The cells were seeded into 96-well plates at a cell density of 1.0 × 104 cells/well in 100 µL of culture medium and maintained for 24 h. Then, the inhibitors with various working concentrations (7.8, 15.6, 31.25, 62.5, 125, 250, and 500 µg/mL) were added to the 96-well plates and incubated for another 48 h. Three wells containing only cells were suspended in a mixture solution of 98 µL complete medium, as the control for cell viability without the inhibitor. Three wells containing only the complete medium were used as blank control. Next, the medium was removed, and 100 µL of fresh culture medium and 10 µL of MTT were added to each well. After incubation for 4 h, the trays were then vigorously shaken to solubilize the formed product, and the absorbance at a wavelength of 490 nm was read on a microplate reader and analyzed. All experiments were conducted in triplicate, and the data are expressed as mean ± standard deviation.
4. Results and Discussion
4.1. UV-Vis Monitoring of Antibiotics Hydrolysis
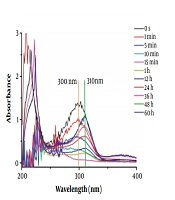
To investigate the inhibition component of carbapenem hydrolysates, initially, the hydrolysis of imipenem and faropenem was monitored by UV-Vis on an Agilent UV8453 UV-Vis spectrometer in a wavelength range of 200 - 600 nm at 25°C, as previously reported (17, 18). The enzymatic and antibiotic samples were prepared with 50 mM Tris buffer, pH = 7.0. In a standard mode, 0.2 µM enzyme solution was used as control. Besides, 10 µL of imipenem (12 mM) or faropenem (8 mM) solution was added into the sample cell filled with 990 µL of ImiS enzyme solution, and the hydrolysis of the antibiotic was monitored until the characteristic absorption of the hydrolysates of the antibiotic had no changes in UV-Vis. The collected UV-Vis spectra are shown in Figure 1. It is clear that imipenem was hydrolyzed quickly in around 50 s, and the maximum absorption of the antibiotic at 300 nm disappeared (Figure 1A). With monitoring for about 1 h, a species with maximum absorption at 310 nm appeared, and with monitoring up to about 48 h, the absorbance of the species reached its maximum.
The monitored UV-Vis spectra of 120 µM imipenem (A) and 80 µM faropenem (B) hydrolyses in the presence of 0.2 µM purified ImiS. Both antibiotic and enzymatic samples were prepared with 50 mM Tris buffer, pH = 7.0.
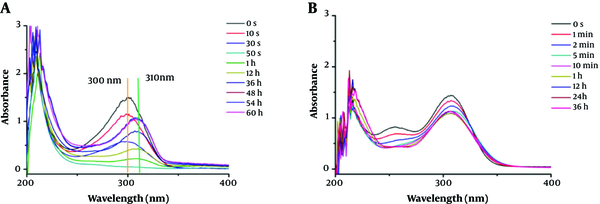
Under the same test conditions, the UV-Vis spectra monitoring of faropenem hydrolysis with the ImiS enzyme is shown in Figure 1B. It shows that faropenem was slowly (compared to imipenem) but surely hydrolyzed, in about 5 min. The difference in the hydrolysis rate was due to the structural differences between the two carbapenems (Appendix 1 in Supplementary File). However, further monitoring, up to 36 h, showed that the absorption of faropenem hydrolysate at 300 nm had no more decreases. Also, the hydrolysate had no absorption at any other wavelengths, except for 300 nm, unlike the hydrolysate of imipenem. This suggests that the hydrolysate was ring-opened faropenem, while the hydrolysate of imipenem was not a ring-opened product.
To investigate the hydrolysate of carbapenems by ImiS inside bacterial cells, the hydrolysis of imipenem and faropenem by E. coli cells with ImiS (E. coli with ImiS) was monitored by UV-Vis (Figure 2). The concentrations of the two antibiotics (imipenen and faropenem) were 120 and 80 µM, respectively, and E. coli with ImiS with an OD600 of 0.1 was used for the evaluation. As expected, imipenem was completely hydrolyzed in E. coli cells, corresponding to a new absorption peak at 310 nm (Figure 2A), in around 15 min, which was longer than the time (5 min) of antibiotic hydrolysis by the ImiS enzyme. This difference may be due to a certain time required for the diffusion of the substrate across the bacterial outer membrane into the periplasm where ImiS was located (20). Meanwhile, the hydrolysis of faropenem by E. coli with ImiS was also monitored (Figure 2B). It can be observed that the antibiotic initially was hydrolyzed in around 20 min, corresponding to the absorption peak at 300 nm, just like the hydrolysis with the ImiS enzyme (Figure 1B), and the absorbance at 300 nm remained stable for 20 min. The imipenem-generated product was distinct from the faropenem-generated product; therefore, the hydrolysates of imipenem with the ImiS enzyme were purified, and their inhibitory activities against MβLs were tested.
The monitored UV-Vis spectra of 120 µM imipenem (A) and 80 µM faropenem (B) hydrolysis in the presence of E. coli cells (OD600 = 0.10) expressing ImiS. Both antibiotic and Escherichia coli samples were prepared with 50 mM Tris buffer, pH = 7.0.
4.2. Inhibitory Activity Evaluation of Active Competent
To ascertain whether the ring-opened product, hydrolyzed carbapenem, was the resulting hydrolyzed and active component, we hydrolyzed imipenem with ImiS, and the resulting hydrolysate was analyzed. A saturated aqueous solution of imipenem was hydrolyzed with ImiS for 60 h at 4°C (it was observed that the solution gradually changed from colorless to yellowish-brown). Then, ImiS was removed from the solution by ultrafiltration (molecular weight cutoff at 10 KD), and the solution was lyophilized overnight to yield a brown powder. The powder was purified on an LC-2010 CHT high-pressure liquid chromatography (HPLC) equipped with a Prep C18 column (10 × 150 mm, 5 µM particle size; Waters), using a gradient running from 5% to 10% water over 20 min with a flow-rate of 1 mL/min, to give one component (P1).
To identify whether P1 was the active competent, the inhibitory activity of P1 was tested against the purified MβLs from different subclasses by UV-Vis via the determination of IC50 values (inhibitory concentration causing 50% decrease in enzyme activity). It was observed that MβLs VIM-2, NDM-1, ImiS, and L1 from B1, B2, and B3 sub-classes were overexpressed and purified as previously described (21-24), respectively (see the Supporting Information). The concentration of P1 ranged from 1 to 50 µM, and the concentrations of imipenem, cefazolin, and nitrocefin (25) as substrates were 40 µM. The enzyme and the inhibitor were pre-incubated for 20 min before adding the substrate and were then monitored at 300, 265, or 390 nm, in sequence, to determine the initial velocity.
The initial reaction rates were determined in triplicate, and the average value was recorded. We tested different pre-incubation times (10 - 120 min) and did not find any difference in residual activity, concluding that P1 is a time-independent inhibitor. The results summarized in Table 1 indicate that P1 exhibited a broad-spectrum inhibition on all enzymes tested (except for ImiS using imipenem as substrate), with an IC50 value in the range of 8 - 32, 13.8 - 29.3, and 14.2 - 19.2 µM, using imipenem, cefazolin, and nitrocefin as substrates, respectively, and the standard deviations of IC50 values were less than 0.17.
Inhibitory Activity (IC50, μM) of Component P1 from the Imipenem Hydrolysate Against Metallo-β-lactamases (MβLs) from Three Sub-Classes in 50 mM Tris, pH = 7.0a, b
| Imipenem | Cefazolin | Nitrocefin | |
|---|---|---|---|
| VIM-2 | 17.8 ± 0.2 | 21.5 ± 0.2 | 17.4 ± 0.3 |
| NDM-1 | 32.0 ± 0.1 | 29.3 ± 0.2 | 19.2 ± 0.1 |
| ImiS | 8.0 ± 0.3 | - | - |
| L1 | 15.3 ± 0.1 | 13.8 ± 0.1 | 14.2 ± 0.2 |
4.3. Structure Determination of P1
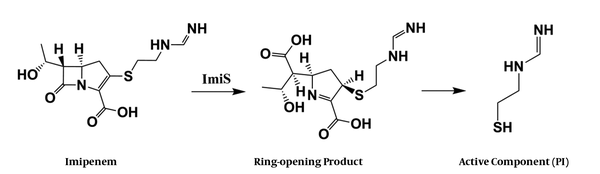
To characterize the active molecule, a sample of 10 mg P1 in 500 µL of D2O was run through 1H and 1H-13C COSY NMR (600 MHz, Bruker), suggesting that P1 could be a mercaptoethyl-form imidamide. As shown in the Appendix 1 in Supplementary File, the single peak at 7.95 ppm is assigned to a proton of an imine, and the two triple split peaks at 3.47 and 2.76 ppm are assigned to two methylene protons. Further, the 1H-13C COSY spectra showed that there are cross-peaks of hydrogen directly bonding to each carbon. Each cross-peak occurs at the point where each carbon atom intersects the chemical shift of the hydrogen on that carbon. Based on the information from the above characterizations, we proposed a hydrolysis process of imipenem by ImiS, as shown in Figure 3, in which P1 is a degradation product of the ring-opened imipenem.
The proposed hydrolysis process of imipenem with ImiS.
To check whether the active molecule has sulfhydryl, P1 was tested with Ellman's reagent (26). A series of solutions of P1, with concentrations ranging from 12.5 - 100 µM, were incubated with 1 mM of Ellman's reagent for 5 min, to determine the absorbance of TNB (2-nitro-5-thiobenzoic acid) at 412 nm. It can be observed (Appendix 1 in Supplementary File) that with the increase of P1 concentration, the solution turned yellow, and the corresponding absorbance of TNB increased linearly from 0.1 to 0.69, confirming that P1 does contain a sulfhydryl, which differs from the product of imipenem hydrolysis by class D β-lactamases (Appendix 1 in Supplementary File) (15). The assays suggested that P1 exists in the mercaptan form under the above activity test conditions.
4.4. Docking Studies
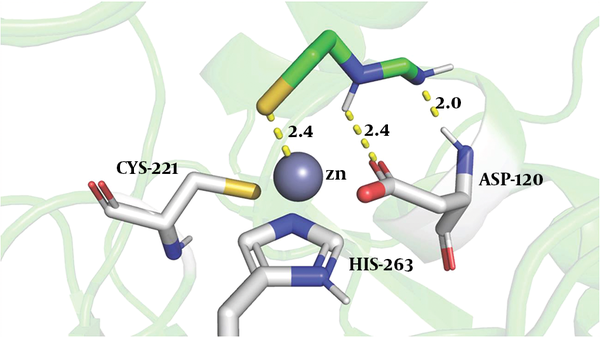
To explore the binding of the imipenem hydrolysate to ImiS, we performed molecular docking of inhibitor P1 into the active sites of CphA (instead of ImiS, which has not been crystallized yet, and with which it shares 96% sequence identity), using the same procedure as reported previously (27). The possible reasonable conformations (shown in Figure 4) are the lowest-energy conformations of those clusters. The sulfur atom of P1 was bound to Zn (II) ion (2.4 Å), the nitrogen atoms of imine and secondary amine formed two hydrogen bonds with carboxylate oxygen and the amino group of Asp120 (2.4 and 2.0 Å), tightly anchoring the inhibitor in the active site, revealing why P1 exhibited a low IC50 value against ImiS.
The conformations of P1 docked into the active site of CphA (PDB code 2QDS). The enzyme backbone is shown as a green cartoon and selected residues are shown as sticks, colored by atom (C, green; N, blue; O, red; S, yellow). The Zn (II) ion is shown as a gray sphere. The key residues displayed and interactions between the P1 and protein residues are indicated by dashed lines.
4.5. Minimum Inhibitory Concentration Determination
Given the broad-spectrum inhibition of imipenem hydrolysate against MβLs, the antibacterial activity of P1 was investigated by determining MIC values of β-lactam antibiotics in the presence and absence of P1. Four bacterial strains, E. coli-DH10B cells harboring plasmids pBCSK(+)-VIM-2, pBCSK(+)-NDM-1, pBCSK(+)-ImiS, and pBCSK(+)-L1, were used in the assays. The antibiotics imipenem and cefazolin (except for the test with E. coli with ImiS, because ImiS exclusively hydrolyzes carbapenems) were used for the assays. The MIC data (Table 2) indicated that a dose of 40 µg/mL of the degradation product P1 restored, by 16 - 32 folds and 8 - 128 folds, the antibacterial activities of imipenem and cefazolin against drug-resistant E. coli with MβLs, respectively. Additionally, P1 exhibited the best synergistic antibacterial efficacy against E. coli with ImiS and L1, resulting in a 32-128-fold decrease in the MIC of these two antibiotics, which is consistent with its inhibitory activity on the ImiS and L1 enzymes, as shown in Table 1. This assay revealed a means for the development of MβLs inhibitors, or even antibacterial reagents, from the hydrolysates of carbapenems.
Antibacterial Activities (MICs, µ/mL) of Imipenem and Cefazolin Against Escherichia coli-DH10B with MβLs in the Absence and Presence of the Degradation Product, P1, of Imipenem Hydrolysate with ImiSa
| Imipenem Alone | Imipenem + P1 | Cefazolin Alone | Cefazolin + P1 | |
|---|---|---|---|---|
| Escherichia coli with VIM-2 | 64 | 4 | 512 | 16 |
| E. coli with NDM-1 | 64 | 4 | 512 | 64 |
| E. coli with ImiS | 64 | 2 | - | - |
| E. coli with L1 | 64 | 2 | 512 | 4 |
4.6. Cytotoxicity Evaluation of P1
The potential toxicity of enzymatic inhibitors is a major concern for their biomedical applications. To determine the maximum non-toxic concentrations of the product inhibitor against mammalian cells, different concentrations of P1 (7.8 - 500 µg/mL) were incubated with L929 mouse fibroblast cells, and cell viability was evaluated using the MTT reagent. The results were expressed as the percent viability of each sample, compared with that of untreated control cells (PBS, pH = 7.4). The cell viability was over 94% in the presence of P1 at a dose up to 500 µM, indicating that the product inhibitor had little influence on cell proliferation (Appendix 1 in Supplementary File).
5. Conclusions
Drug-resistant bacterial infections caused by MβLs have grown into an emerging threat, where screening the inhibitors, specifically, the broad-spectrum inhibitors of MβLs, has proven to be challenging. In this work, we monitored the hydrolysis of imipenem and faropenem by ImiS and found that the hydrolysate of imipenem exhibited absorption at 310 nm, which is different from that of the antibiotic (300 nm), and the hydrolysate was separated by HPLC to give one component P1. The kinetic assays revealed that P1 exhibited a broad-spectrum inhibition on tested VIM-2, NDM-1, ImiS, and L1, from three sub-classes of MβLs, with an IC50 value ranging from 8 - 32, 13.8 - 29.3, and 14.2 - 19.2 µM using imipenem, cefazolin, and nitrocefin as substrates, respectively. Further, P1 showed synergistic antibacterial efficacy against drug-resistant E. coli with MβLs, restoring by 16-32-fold and 32-128-fold activities of imipenem and cefazolin, respectively.
Spectroscopic characterization and Ellman's reagent analysis suggested that P1, a mercaptoethyl-form imidamide from the leaving group produced after imipenem is hydrolyzed, is a mechanism-based inhibitor, while faropenem has no substrate inhibition, probably due to the lack of a good leaving group. Docking studies revealed that the sulfur atom of P1 was bound to the Zn (II) ion at the active site, and the nitrogen atoms of the imine and secondary amine formed two hydrogen bonds with the carboxylate oxygen and amino groups of Asp120. Toxicity tests indicated that P1 had little influence on cell proliferation at a dose up to 500 µM. This work reveals that the hydrolysates of carbapenems with good leaving groups can be useful in screening for broad-spectrum inhibitors of MβLs.
References
-
1.
Ventola CL. The antibiotic resistance crisis: part 1: Causes and threats. P T. 2015;40(4):277-83. [PubMed ID: 25859123]. [PubMed Central ID: PMC4378521].
-
2.
Lassaux P, Hamel M, Gulea M, Delbruck H, Mercuri PS, Horsfall L, et al. Mercaptophosphonate compounds as broad-spectrum inhibitors of the metallo-beta-lactamases. J Med Chem. 2010;53(13):4862-76. [PubMed ID: 20527888]. https://doi.org/10.1021/jm100213c.
-
3.
Crowder MW, Spencer J, Vila AJ. Metallo-beta-lactamases: Novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39(10):721-8. [PubMed ID: 17042472]. https://doi.org/10.1021/ar0400241.
-
4.
Heinz U, Adolph HW. Metallo-beta-lactamases: Two binding sites for one catalytic metal ion? Cell Mol Life Sci. 2004;61(22):2827-39. [PubMed ID: 15558212]. https://doi.org/10.1007/s00018-004-4214-9.
-
5.
Hernandez Valladares M, Felici A, Weber G, Adolph HW, Zeppezauer M, Rossolini GM, et al. Zn(II) dependence of the Aeromonas hydrophila AE036 metallo-beta-lactamase activity and stability. Biochemistry. 1997;36(38):11534-41. [PubMed ID: 9298974]. https://doi.org/10.1021/bi971056h.
-
6.
Mohamed MS, Hussein WM, McGeary RP, Vella P, Schenk G, Abd El-Hameed RH. Synthesis and kinetic testing of new inhibitors for a metallo-beta-lactamase from Klebsiella pneumonia and Pseudomonas aeruginosa. Eur J Med Chem. 2011;46(12):6075-82. [PubMed ID: 22051063]. https://doi.org/10.1016/j.ejmech.2011.10.030.
-
7.
Toney JH, Hammond GG, Fitzgerald PM, Sharma N, Balkovec JM, Rouen GP, et al. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-beta-lactamase. J Biol Chem. 2001;276(34):31913-8. [PubMed ID: 11390410]. https://doi.org/10.1074/jbc.M104742200.
-
8.
Toney JH, Fitzgerald PM, Grover-Sharma N, Olson SH, May WJ, Sundelof JG, et al. Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase. Chem Biol. 1998;5(4):185-96. [PubMed ID: 9545432]. https://doi.org/10.1016/s1074-5521(98)90632-9.
-
9.
Chiou J, Wan S, Chan KF, So PK, He D, Chan EW, et al. Ebselen as a potent covalent inhibitor of New Delhi metallo-beta-lactamase (NDM-1). Chem Commun (Camb). 2015;51(46):9543-6. [PubMed ID: 25970101]. https://doi.org/10.1039/c5cc02594j.
-
10.
Rossolini GM, Walsh T, Amicosante G. The Aeromonas metallo-beta-lactamases: genetics, enzymology, and contribution to drug resistance. Microb Drug Resist. 1996;2(2):245-52. [PubMed ID: 9158767]. https://doi.org/10.1089/mdr.1996.2.245.
-
11.
Kramer DM. Photosynth research. Photosynth Res. 2000;6:291-2.
-
12.
Sharma N, Hu Z, Crowder MW, Bennett B. Conformational changes in the metallo-beta-lactamase ImiS during the catalytic reaction: An EPR spectrokinetic study of Co(II)-spin label interactions. J Am Chem Soc. 2008;130(26):8215-22. [PubMed ID: 18528987]. [PubMed Central ID: PMC2574873]. https://doi.org/10.1021/ja0774562.
-
13.
Costello AL, Sharma NP, Yang KW, Crowder MW, Tierney DL. X-ray absorption spectroscopy of the zinc-binding sites in the class B2 metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry. 2006;45(45):13650-8. [PubMed ID: 17087519]. https://doi.org/10.1021/bi061547e.
-
14.
Crawford PA, Yang KW, Sharma N, Bennett B, Crowder MW. Spectroscopic studies on cobalt(II)-substituted metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry. 2005;44(13):5168-76. [PubMed ID: 15794654]. https://doi.org/10.1021/bi047463s.
-
15.
Lohans CT, van Groesen E, Kumar K, Tooke CL, Spencer J, Paton RS, et al. A new mechanism for beta-lactamases: Class D enzymes degrade 1beta-methyl carbapenems through lactone formation. Angew Chem Int Ed Engl. 2018;57(5):1282-5. [PubMed ID: 29236332]. [PubMed Central ID: PMC5817396]. https://doi.org/10.1002/anie.201711308.
-
16.
Sharma NP, Hajdin C, Chandrasekar S, Bennett B, Yang KW, Crowder MW. Mechanistic studies on the mononuclear ZnII-containing metallo-beta-lactamase ImiS from Aeromonas sobria. Biochemistry. 2006;45(35):10729-38. [PubMed ID: 16939225]. [PubMed Central ID: PMC2597473]. https://doi.org/10.1021/bi060893t.
-
17.
Yang KW, Zhou Y, Ge Y, Zhang Y. Real-time activity monitoring of New Delhi metallo-beta-lactamase-1 in living bacterial cells by UV-Vis spectroscopy. Chem Commun (Camb). 2017;53(57):8014-7. [PubMed ID: 28664213]. https://doi.org/10.1039/c7cc02774e.
-
18.
Ge Y, Zhou YJ, Yang KW, Zhang YL, Xiang Y, Zhang YJ. Real-time activity assays of beta-lactamases in living bacterial cells: Application to the inhibition of antibiotic-resistant E. coli strains. Mol Biosyst. 2017;13(11):2323-7. [PubMed ID: 28906528]. https://doi.org/10.1039/c7mb00487g.
-
19.
Sader HS, Flamm RK, Jones RN. Antimicrobial activity of daptomycin tested against Gram-positive pathogens collected in Europe, Latin America, and selected countries in the Asia-Pacific Region (2011). Diagn Microbiol Infect Dis. 2013;75(4):417-22. [PubMed ID: 23514757]. https://doi.org/10.1016/j.diagmicrobio.2013.01.001.
-
20.
Zhang YJ, Wang WM, Oelschlaeger P, Chen C, Lei JE, Lv M, et al. Real-time monitoring of NDM-1 activity in live bacterial cells by isothermal titration calorimetry: A new approach to measure inhibition of antibiotic-resistant bacteria. ACS Infect Dis. 2018;4(12):1671-8. [PubMed ID: 30383355]. https://doi.org/10.1021/acsinfecdis.8b00147.
-
21.
Aitha M, Marts AR, Bergstrom A, Moller AJ, Moritz L, Turner L, et al. Biochemical, mechanistic, and spectroscopic characterization of metallo-beta-lactamase VIM-2. Biochemistry. 2014;53(46):7321-31. [PubMed ID: 25356958]. [PubMed Central ID: PMC4245990]. https://doi.org/10.1021/bi500916y.
-
22.
Yang H, Aitha M, Hetrick AM, Richmond TK, Tierney DL, Crowder MW. Mechanistic and spectroscopic studies of metallo-beta-lactamase NDM-1. Biochemistry. 2012;51(18):3839-47. [PubMed ID: 22482529]. https://doi.org/10.1021/bi300056y.
-
23.
Crawford PA, Sharma N, Chandrasekar S, Sigdel T, Walsh TR, Spencer J, et al. Over-expression, purification, and characterization of metallo-beta-lactamase ImiS from Aeromonas veronii bv. sobria. Protein Expr Purif. 2004;36(2):272-9. [PubMed ID: 15249050]. https://doi.org/10.1016/j.pep.2004.04.017.
-
24.
Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Overexpression, purification, and characterization of the cloned metallo-beta-lactamase L1 from Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1998;42(4):921-6. [PubMed ID: 9559809]. [PubMed Central ID: PMC105568]. https://doi.org/10.1128/AAC.42.4.921.
-
25.
van Berkel SS, Brem J, Rydzik AM, Salimraj R, Cain R, Verma A, et al. Assay platform for clinically relevant metallo-beta-lactamases. J Med Chem. 2013;56(17):6945-53. [PubMed ID: 23898798]. [PubMed Central ID: PMC3910272]. https://doi.org/10.1021/jm400769b.
-
26.
Riddles PW, Blakeley RL, Zerner B. Ellman's reagent: 5,5′-dithiobis(2-nitrobenzoic acid)-a reexamination. Anal Biochem. 1979;94(1):75-81. https://doi.org/10.1016/0003-2697(79)90792-9.
-
27.
Zhang YL, Yang KW, Zhou YJ, LaCuran AE, Oelschlaeger P, Crowder MW. Diaryl-substituted azolylthioacetamides: Inhibitor discovery of New Delhi metallo-beta-lactamase-1 (NDM-1). Chem Med Chem. 2014;9(11):2445-8. [PubMed ID: 25048031]. https://doi.org/10.1002/cmdc.201402249.