Keywords
Dear editor,
Direct fungal DNA extraction using commercial kits from whole blood or its components, such as serum and plasma, is challenging with poor sensitivity, mainly due to the abundance of red blood cells and the low number of fungal cells present. Fungal DNA extraction from positive fungal blood culture bottles offers a higher chance of DNA recovery for detection as the number of fungal cells increases. However, this extraction is often challenging due to many inhibitory factors, i.e., abundant interfering human DNA, blood preservatives, and rich blood cell components (1). Thus, we proposed a modified extraction method that successfully detected and identified various Candida species to avoid problems mentioned above. This method helps shorten the relative turn-around-time, TAT (pure culture is usually used for DNA extraction in normal circumstances), and avoid using toxic chemicals, i.e. phenol-chloroform, lysis buffer, proteinase K, or expensive lyticase enzymes. It overcomes the limitation of existing commercial kits usually designed either for blood or fungal DNA extraction (2, 3).
We developed the in-house method that involved incubating 1 ml of positive blood culture bottle sample in a heating block at 95°C for 30 min with vortexing for 15 s every 10 min. It was followed by centrifugation at 18,000 g for 10 min. Next, the supernatant was transferred to a sterile microcentrifuge tube with an equal volume of absolute ethanol. A 1/10th volume of 3M sodium acetate, pH 5.2, was gently added and mixed by inversion to precipitate the DNA and incubated on ice for 20 min. Following incubation, the tube was centrifuged for 15 min at 11,000 x g to obtain the pellet. The pellet containing the extracted DNA with minute blood traces (haem) was purified using the ExpinTM Combo GP kit (GeneAll Biotechnology, South Korea). The DNA eluates were kept at -20°C until use, as indicated in the schematic illustration (Figure 1A). Standard PCR assay with initial denaturation: 95°C, 4 min; denaturation: 95°C, 20 s; annealing: 62°C, 15 s; and extension: 72°C, 30 s, with one cycle for initial denaturation and 30 cycles repeated for denaturation, annealing, and extension using universal pan-fungal internal transcribed spacer (ITS) primers successfully amplified the ITS region of the extracted DNA samples (Figure 1B).
A, modified DNA extraction protocol for Candida isolates; B, duplicate PCR results. Legend: Lane M, DNA molecular weight marker 100 bp (ThermoFisher); Lanes NC, no template control; Lanes 1 & 8, C. glabrata; Lanes 2 & 9: C. tropicalis; Lanes 3 & 10: C. albicans; Lanes 4 & 11, C. tropicalis; Lanes 5 & 12, C. albicans; Lanes 6 & 13, C. auris; Lane PC, positive control (C. glabrata ATCC 2001) (~ 848 bp).
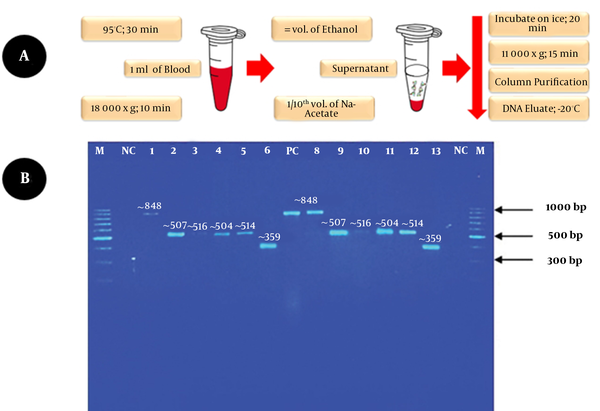
The identification of the isolates was confirmed by direct sequencing of the purified PCR products. The interpreted sequencing results identified six clinical Candida species of different amplicon sizes between 350 - 850 bp. The identified species included C. glabrata (~ 848 bp), C. tropicalis (~ 507 bp), C. albicans (~ 516 bp), C. tropicalis (~ 504 bp), C. albicans (~ 516 bp), and C. auris (~ 359 bp) (Figure 1B). The extracted DNA showed good integrity and shortened the TAT. The yield is adequate as a template for PCR and simple cloning. The method is simple, rapid, affordable, and environmentally friendly. It helps rapid detection of bloodstream yeast Candida species from positive blood culture bottles. Hence, it can help initiate timely and appropriate antifungal therapy
References
-
1.
Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36(2):288-305. [PubMed ID: 21569057]. https://doi.org/10.1111/j.1574-6976.2011.00278.x.
-
2.
Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27(3):490-526. [PubMed ID: 24982319]. [PubMed Central ID: PMC4135902]. https://doi.org/10.1128/CMR.00091-13.
-
3.
Griffiths L, Chacon-Cortes D. Methods for extracting genomic DNA from whole blood samples: Current perspectives. J Biorepository Sci Appl Med. 2014;2:1-9. https://doi.org/10.2147/bsam.s46573.