Abstract
Background:
Enterococci are one of the opportunistic pathogenic microorganisms that can cause significant problems for human and animal health. Enterococcus faecium seems to be more resistant to antibiotics than E. faecalis. It is thought that pathogenic E. faecium can develop antibiotic resistance very quickly, and the ability to transfer this feature is considered to be an important health risk.Objectives:
This study aimed to determine the prevalence, biotypes, and in vitro antimicrobial susceptibility of E. faecalis and E. faecium strains isolated from 267 routine urine and stool samples that were brought to the microbiology laboratory of Regional Training and Research Hospital of Van, with permission of the patients.Methods:
In the present study, enterococci using species-specific primers to examine E. faecalis and E. faecium multiplex PCR technique was applied. Biotyping of the isolates was used to identify them as E. faecalis and E. faecium by molecular techniques, and antibiotic susceptibility of all samples was examined, as well.Results:
The isolates were identified by multiplex PCR using species-specific primers for E. faecalis and E. faecium. Biotyping based on 13 biochemical tests showed that 72.5%, 12.5%, and 15% of E. faecalis strains were of biotypes I, II, and III, respectively, whereas E. faecium strains could be divided into biotype I (10%), biotype II (12.5%), biotype III (27.5%), and biotype IV (50%). Additionally, all E. faecalis strains were found to be susceptible to penicillin G and imipenem. On the other hand, 95% of the E. faecalis strains were found to be resistant to clindamycin, 77.5% to tetracycline and trimethoprim/sulfamethoxazole, 42.5% to erythromycin, 32.5% to gentamicin, and 17.5% to ciprofloxacin. Of E. faecium strains, 37.5% were found to be resistant to clindamycin, 32.5% to penicillin G, 27.5% to erythromycin and imipenem, 20% to ciprofloxacin, 17.5% to tetracycline and trimethoprim/sulfamethoxazole, 15% to gentamicin, and 5% to vancomycin.Conclusions:
In conclusion, the identification of E. faecalis and E. faecium strains by PCR is reliable and faster than biochemical tests. Additionally, the results of antimicrobial susceptibility tests may provide important contributions to the clinical approach.Keywords
Antimicrobial Susceptibility Biotyping Enterococcus faecium E. faecalis
1. Background
Enterococcus spp. are Gram-positive, facultative, anaerobic bacteria and are present as part of the commensal bacteria in the intestinal tracts of humans and animals (1, 2). They are not considered to be pathogenic (3). Some of these species are also known as the causes of causal diseases in immunocompromised hosts (4). Bacteria of the genus Enterococcus are the components of the natural microbiota of the gastrointestinal tract in both humans and animals. On the other hand, enterococci are considered opportunistic pathogens mainly responsible for nosocomial infections in humans (5), as well as many types of infections in animals such as mastitis in cattle, diarrhea in swine and cattle, and septicaemic diseases in poultry (6).
Their ubiquitous nature, resistance to unfavorable environmental factors, and the capability of the acquisition of antimicrobial resistance and virulence factors also have contributed to the increased share of Enterococcus in opportunistic infections (5, 7). Furthermore, they have now become one of the most common nosocomial infections and are also an emerging threat to public health, as they are intrinsically resistant to several antimicrobials (8-10). These species also show different patterns of antimicrobial resistance, possibly caused by various mechanisms of resistance among them (11). Enterococcus faecium and E. faecalis are major species associated with several enterococcal diseases. Enterococcus faecalis and E. faecium can produce potential virulence factors that may enhance their pathogenicity. In other words, they are responsible for diseases (3, 12). Moreover, E. faecalis and E. faecium have a highly developed ability to acquire resistance genes from the same or different species via transferable plasmids or transposons (13).
Enterococci now account for about 10% of hospital-acquired bacteremia cases globally, and they are the fourth and fifth leading causes of sepsis in North America and Europe, respectively (14). Among the medically important enterococci, E. faecium, in particular, has become a leading cause of nosocomial infections. Enterococcus faecium population analysis has revealed the emergence of a rapidly evolving lineage referred to as Clade-A1 that includes clonal complex 17 (CC17), comprising strains associated with hospital infections across five continents. These hospital strains are resistant to ampicillin, aminoglycosides, and quinolones, and their genomes contain a high number of mobile genetic elements and are enriched for genes encoding altered carbohydrate utilization and transporter proteins that distinguish them from community-acquired and nonpathogenic E. faecium strains (15).
2. Objectives
In this study, we aimed to demonstrate the biotype profiles of E. faecalis and E. faecium strains isolated from stool and urine samples of humans and determine their in vitro susceptibility to various antibiotics by the disk diffusion method. It is thought that the results can be a source for the diagnosis and treatment protocols in diseases caused by these factors and give an idea about the steps of resistance.
3. Methods
3.1. Urine and Stool Samples
In this study, we used routine urine and stool samples brought to the microbiology laboratory of the Van Regional Training and Research Hospital for the diagnosis of urinary tract or digestive system infections.
3.2. Isolated Bacteria and Reference Strains
In this study, E. faecalis and E. faecium strains were isolated and identified in urine and stool samples. Enterococcus faecalis ATCC 29212, E. faecalis ATCC 51299, and E. faecium ATCC 19434 reference strains were used for quality control in molecular diagnosis, biochemical tests, and antimicrobial susceptibility tests.
3.3. Bacterial Isolation
For the isolation of E. faecalis and E. faecium strains, blood agar, Edwards medium, and even Esculin agar were cultured with the help of sterile swabs from urine and stool samples, which had been thoroughly homogenized. The incubation was done at 37°C for 24 - 48 hours under aerobic conditions. The growth of S-type Esculin-positive black colonies was evaluated (16).
3.4. DNA Isolation
The DNA isolation of 24 h bacterial cultures separated as positive controls, and suspicious enterococci was performed using a DNA isolation kit (Thermo GeneJET Genomic DNA Purification Kit - K0722 Lithuania).
3.5. Primer Design
Primers were verified and designed with the GenBank database based on specific regions of DNA encoding 16S and/or 23S rRNA genes. Primer sequences were F: 5 ‘ACT TAT GTG ACT AAC TTA ACC 3’ and R: 5 ‘TAA TGG TGA ATC TTG GTT TGG 3’ for E. faecalis and F: 5 ‘GAA AAA ACA ATA GAA GAA TTA T 3’ and R: 5 ‘TGC TTT TTT GAA TTC TTC TTT A 3’ for E. faecium (17).
3.6. Amplification
The multiplex PCR technique was applied as reported. Master mixes (Thermo PCR Mastermix 2x – K0171 Lithuania) were used for the amplification step. For this purpose, 2 µL bacterial DNA, 1 µL F and R primers, 25 µL mastermix were pooled and the mixture was completed to 50 µL with PCR grade water. The PCR condition was as follows: Pre-denaturation for 10 min at 96°C, followed by 35 cycles of denaturation at 95°C for 45 s, binding at 49°C for 45 s, elongation at 72°C for 60 s, and final elongation at 72°C for 10 s protocol was applied (17, 18).
3.7. Agarose Gel Electrophoresis
The products obtained at the end of the amplification stage were run on a 1% agarose gel with positive controls and examined in the imaging system (Genesis). While evaluating the bands, it was investigated whether the isolates that were E. faecalis-positive and the isolates that were E. faecium-positive formed the bands of 360 bp and 215 bp, respectively.
3.8. Biotyping
Despite the known biochemical characteristics of enterococci, variations can be observed in the biochemical features of many field strains. These variations can cause problems and may lead to wrong identification. Therefore, it holds great importance to determine the biotyping profiles of strains identified by molecular methods. Biotyping of the isolates identified as E. faecalis and E. faecium by molecular techniques was conducted by hemolysis tests, growth in 6.5% NaCl media, hydrolysis of esculin and arginine, H2S production, indole, motility, fermentation of L-arabinose, arbutin, galactose, trehalose, D-glucose, mannitol, D-raffinose, salicin, lactose and sorbitol tests (16, 19, 20).
3.9. Antibiotic Susceptibility Test
The suscepbility of isolates to penicillin-G (10 IU), vancomycin (30 µg), imipenem (10 µg), ciprofloxacin (5 µg), rifampicin (30 µg), gentamicin (10 µg), tetracycline (30 µg), erythromycin (15 µg), clindamycin (2 µg) and trimethoprim/sulfamethoxazole (SXT) (25 µg) was investigated by using Kirby-Bauer disc diffusion method. The evaluations were performed according to CLSI M02-A11 (M100-S23 - The Application Standards for the Antimicrobial Susceptibility Tests) (21, 22).
3.10. Statistical Analysis
A statistical anlaysis was carried out using the cluster variable method in the Minitab package program (Demo Ver-16).
4. Results
4.1. Bacterial Isolation and Identification
Preliminary identification of Enterococcus spp. was based on phenotypic characteristics, such as esculin positive, brown-black S type colony-forming, Gram-positive, single, double, or chain-shaped catalase-negative cocci. A total of 187 Enterococcus spp. including 119 (67.2%) from 177 urine samples and 68 (75.5%) from 90 stool samples were isolated and identified.
4.2. Identification of Enterococcal Isolates by Multiplex PCR
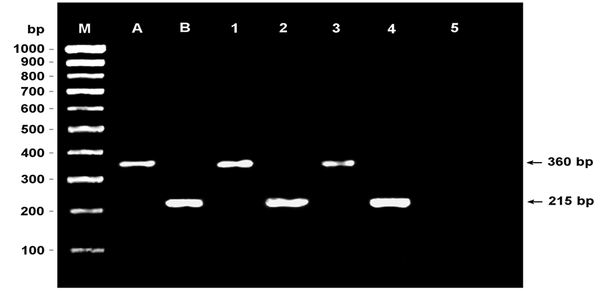
In the identification of isolates using the PCR technique with species-specific primers, 30 (25.2%) of urine originated Enterococcus strains were identified as E. faecalis and eight (6.7%) as E. faecium. Besides, 10 (14.7%) of the stool originated isolates were identified as E. faecalis and 32 (47%) as E. faecium (Figure 1).
Agarose gel image of the analysis of enterococci by the multiplex PCR method (M: PCR ranger 100 bp DNA marker; A: Enterococcus faecalis positive control; B: E. faecium positive control; 1: E. faecalis positive stool specimen; 2: E. faecium positive stool specimen; 3: E. faecalis positive urine specimen; 4: E. faecium positive urine specimen; 5: negative control).
4.3. Biotyping of Enterococcus Strains
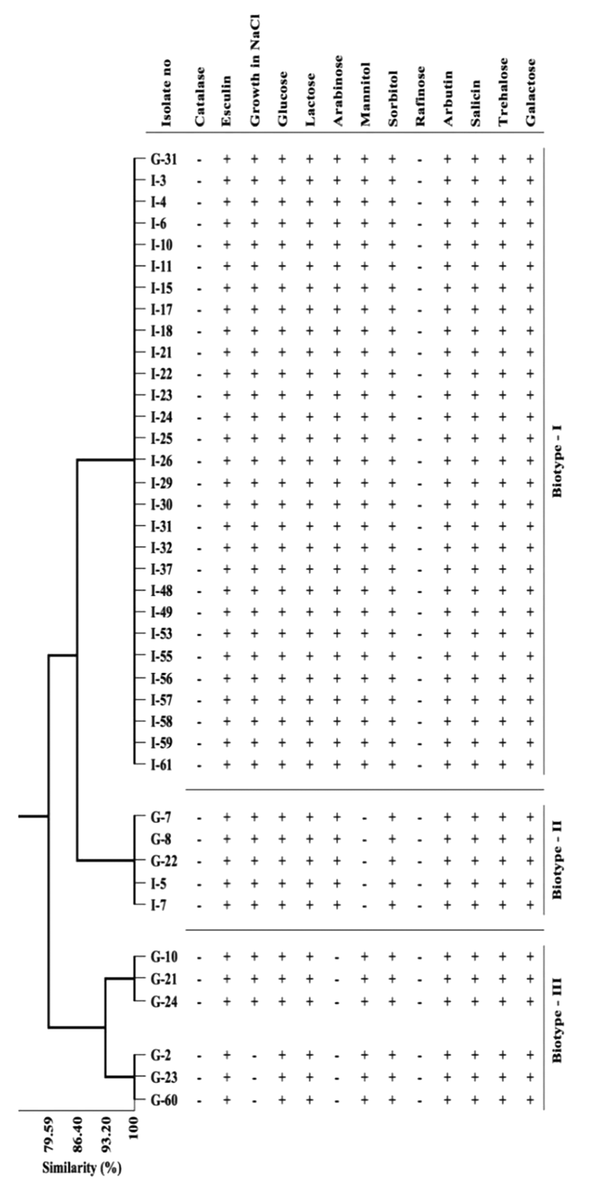
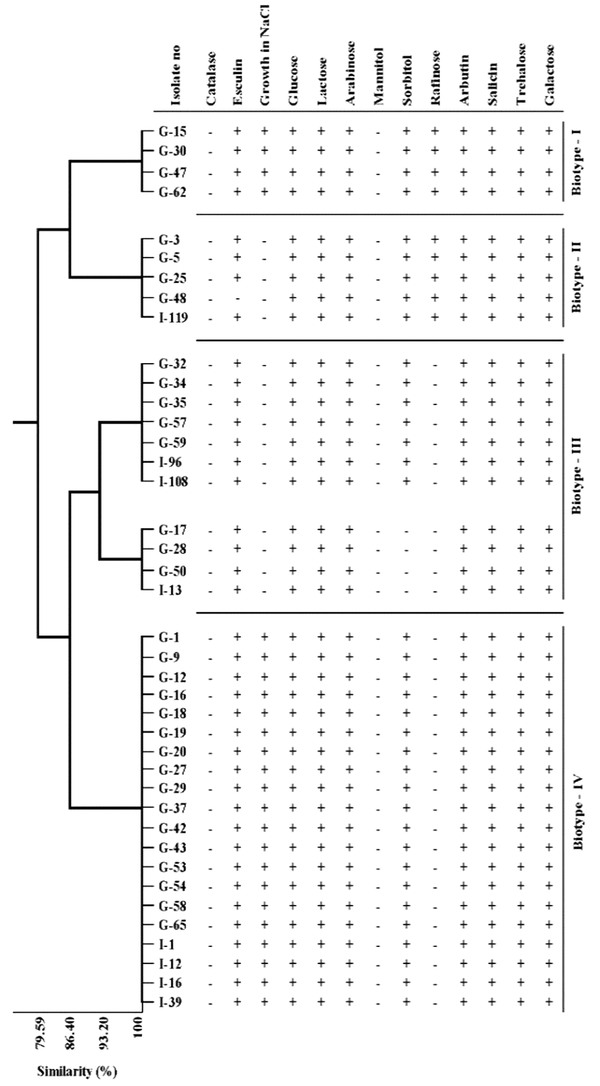
The biochemical test results of E. faecium and E. faecalis isolates that identified by multiplex PCR, were presented in Table 1. While all the strains of E. faecium and E. faecalis were found to be positive for esculin, 45% of E. faecalis isolates and 20% of E. faecium isolates were β-hemolytic. Besides, 92.5% of E. faecalis and 60% of E. faecium cultures were found to be positive for growth in 6.5% NaCl media. The biochemical properties of the isolates were analyzed statistically by using the cluster variable method. Three biotype profiles were found in E. faecalis strains and four biotype profiles in E. faecium strains (Table 2, Figures 2 and 3). Of the E. faecalis strains, 72.5% were determined as biotype I, 12.5% as biotype II, and 15% as biotype III. Moreover, 10% of E. faecium strains were determined as biotype I, 12.5% as biotype II, 27.5% as biotype III, and 50% as biotype IV (Table 2).
Biochemical Test Results of Enterococcus faecalis and E. faecium Isolates
| Tests | Positive Reactions (%) | |
|---|---|---|
| Enterococcus faecalis | Enterococcus faecium | |
| β-hemolysis | 18 (45) | 8 (20) |
| α-hemolysis | 0 | 2 (5) |
| γ-hemolysis | 22 (55) | 30 (75) |
| Growth in 6.5% NaCl | 37 (92.5) | 24 (60) |
| Esculin hydrolysis | 40 (100) | 40 (100) |
| Arginin hydrolysis | 40 (100) | 40 (100) |
| H2S | 0 | 0 |
| Indol | 0 | 0 |
| Motility | 0 | 0 |
| L-Arabinose | 34 (85) | 40 (100) |
| Arbutin | 40 (100) | 40 (100) |
| Galactose | 40 (100) | 40 (100) |
| Glucose | 40 (100) | 40 (100) |
| Lactose | 40 (100) | 40 (100) |
| Mannitol | 35 (87.5) | 0 |
| Raffinose | 0 | 9 (22.5) |
| Salicin | 40 (100) | 40 (100) |
| Sorbitol | 40 (100) | 36 (90) |
| Trehalose | 40 (100) | 40 (100) |
Distribution of Biotype Profiles of Enterococcus faecalis and E. faecium Isolates
| Biotypes | Number of Biotypes (%) | |
|---|---|---|
| E. faecalis | E. faecium | |
| Biotype-I | 29 (72.5) | 4 (10) |
| Biotype-II | 5 (12.5) | 5 (12.5) |
| Biotype-III | 6 (15) | 11 (27.5) |
| Biotype-IV | - | 20 (50) |
Distribution of biotype profiles and similarities of Enterococcus faecalis isolates
Distribution of biotype profiles and similarities of Enterococcus faecium isolates
4.4. Antimicrobial Susceptibility Test Results
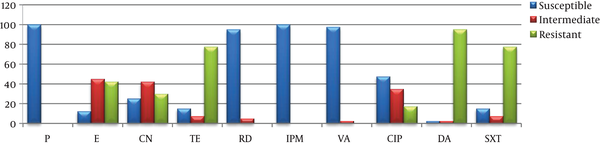
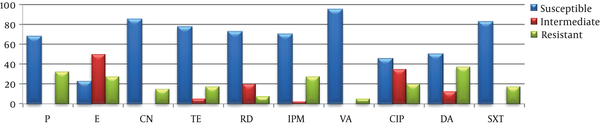
All the E. faecalis strains examined by the disk diffusion method were susceptible to imipenem and penicillin G while no vancomycin-resistant isolate was determined. Of them, 95% were found to be resistant to clindamycin and 77.5% to tetracycline and SXT. Of the studied E. faecium types, 95% were found to be susceptible to vancomycin and 67.5% to penicillin G (Table 3, Figures 4 and 5).
In Vitro Antimicrobial Disk Diffusion Test Results of Enterococcus faecium and E. faecalis Isolates
| Antimicrobial Agent | Enterococcus faecalis (%) | Enterococcus faecium (%) | ||
|---|---|---|---|---|
| Susceptible | Intermediate | Susceptible | Intermediate | |
| Penicillin-G | 40 (100) | 0 | 27 (67.5) | 0 |
| Erythromycin | 5 (12.5) | 18 (45) | 9 (22.5) | 20 (50) |
| Gentamicin | 10 (25) | 17 (42.5) | 34 (85) | 0 |
| Tetracycline | 6 (15) | 3 (7.5) | 31 (77.5) | 2 (5) |
| Rifampicin | 38 (95) | 2 (5) | 29 (72.5) | 8 (20) |
| Imipenem | 40 (100) | 0 | 28 (70) | 1 (2.5) |
| Vancomycin | 39 (97.5) | 1 (2.5) | 38 (95) | 0 |
| Ciprofloxacin | 19 (47.5) | 14 (35) | 18 (45) | 14 (35) |
| Clindamycin | 1 (2.5) | 1 (2.5) | 20 (50) | 5 (12.5) |
| SXT | 6 (15) | 3 (7.5) | 33 (82.5) | 0 |
Distribution of in vitro antimicrobial disk diffusion test results of Enterococcus faecalis isolates
Distribution of in vitro antimicrobial disk diffusion test results of Enterococcus faecium isolates
5. Discussion
In the present study, the isolates were identified by multiplex PCR with species-specific primers for E. faecalis and E. faecium. Out of 187 isolates, 30 (25.2%) and 10 (14.7%) were identified to be E. faecalis in urine and stool samples, respectively, whereas eight (6.7%) isolates from stool samples and 32 (47%) isolates from urine samples were identified as E. faecium. In a study, out of 280 enterococcal isolates, 175 isolates were identified as E. faecalis, 67 as E. faecium, and 38 as Enterococcus spp. (23). In another study, the prevalence of E. faecalis and E. faecium isolates were reported 76% and 55.5%, respectively (24).
Manero and Blanch (25) applied 94 different biochemical tests with referenced Enterococcus cultures and 82 clinical isolates, and 76 of these tests could be used to identify Enterococcus species and 12 of them (D-raffinose, L-arabinose, sorbose, ribose, methyl-α-D-glucopyranoside, mannitol, arginine, sucrose, pyrrolidinyl aminopeptidase, α-galactose, yellow pigment production, and alkaline phosphate) could be used safely for the identification of Enterococcus strains based on species.
Facklam and Collins (26) stated that three different biotypes were identified according to biochemical features of 206 cultures identified as Enterococcus spp. previously; besides, nine different biotype profiles identified by API-20 kits were reported in their study. Pelicioli Riboldi et al. (27) also identified eight different biotypes in 55 Enterococcus strains. In the current study, 3 biotype profiles were identified in E. faecalis strains and 4 biotype profiles were identified in E. faecium strains according to biochemical characteristics of isolates. Out of 40 E. faecalis strains, 72.5% of them were identified as biotype I, 12.5% as biotype II and 15% as biotype III, while 10% of E. faecium strains identified as biotype I, 12.5% biotype II, 27.5% were biotype III and 50% were biotype IV. It was observed that the findings of the study are different from the other studies that unlike one another.
The development of multiple antimicrobial resistance worldwide has begun to cause serious problems, especially by nosocomial Enterococcus strains (28-31). The development of resistance to antibiotics occurs by the acquisition of plasmids or transposons containing resistance genes or by mutations. Despite many studies conducted on antibiotic resistance of Enterococcus species, studies on pathogenicity mechanisms and virulence factors have been inadequate (32).
The antibiotic resistance mechanisms in enterococci can be explained in two ways. Intrinsic resistance is inherently encoded in chromosomes in most or all types of enterococci. The observed structural resistance mechanisms for some antibiotics are typically specific to some types of Enterococcus species. Acquired resistance is more variable than intrinsic resistance. Enterococci are able to develop resistance to many antibiotics by means of plasmids or transposons, by this way, tetracyclines have become resistant to macrolides, lincosamides, and chloramphenicol. They show a high level of aminoglycoside resistance with a large number of different aminoglycosides by modifying enzymes. In addition to all these, the resulting plasmid- based beta-lactam resistance is another problem (1, 5, 28, 33).
In the studies investigating the resistance of E. faecalis strains to antimicrobial agents isolated from various clinical samples, Berzeg et al. (34) found that E. faecalis strains were resistant to imipenem while they were resistant to penicillin at 4%, ciprofloxacin at 8%, rifampicin at 40%, and gentamicin at 8%. Aktepe et al. (35) reported that E. faecalis strains were resistant to imipenem at 51.9%. Aral et al. (36) stated that 27% of E. faecalis strains were resistant to ciprofloxacin, 16% to gentamicin, 56% to erythromycin, and all were resistant to clindamycin and SXT while they were sensitive to imipenem. Iraz et al. (37) reported that E. faecalis strains were resistant to vancomycin at 4%, ciprofloxacin at 47%, and gentamicin at 42%. Altun et al. (38) observed that 16% of E. faecalis strains were resistant to penicillin and 44% to gentamicin, while all were susceptible to vancomycin. Güçkan et al. (39) detected that E. faecalis strains were resistant to ciprofloxacin at 50%, gentamycin at 44%, tetracycline at 68%, clindamycin at 95%, and SXT at 100%. Barisic and Punda Polic (40) found aminoglycoside resistance in 37% of E. faecalis isolates from hospitalized patients. In another study involving 27 European countries, gentamicin resistance was 20%, vancomycin at 0.03%, erythromycin at 47%, imipenem at 1%, and ciprofloxacin resistance was found to be present in 6% of E. faecalis strains (41).
In this study, all the examined E. faecalis isolates were found to be susceptible to penicillin, rifampicin, imipenem, and vancomycin, while 42.5% were resistant to erythromycin, 32.5% to gentamicin, 77.5% to tetracycline, 17.5% to ciprofloxacin, 95% to clindamycin, and 77.5% to SXT. In studies conducted on E. faecium, Berzeg et al. (34) reported that the examined E. faecium types isolated from clinical samples were resistant to penicillin at 68%, imipenem at 27%, and ciprofloxacin, rifampicin, and gentamicin at 68%. Aktepe et al. (35) found that 70% of the strains were resistant to imipenem. Aral et al. (36) stated that 94% of the isolates were resistant to imipenem, 69% to ciprofloxacin, 60% to gentamicin, 99% to clindamycin, and all were resistant to erythromycin and SXT. Iraz et al. (37) reported that 23% of the isolates were resistant to vancomycin, 84% to ciprofloxacin, and 69% to gentamicin. Altun et al. (38) expressed that the resistance rates of the studied cultures were 83% to penicillin, 16.1% to vancomycin, and 71% to gentamicin. Güçkan et al. (39) determined resistance to ciprofloxacin at 44%, gentamicin at 40%, tetracycline at 60%, clindamycin at 89%, and SXT at 98%. In another study involving 27 European countries, gentamicin resistance was 22,5%, vancomycin at 2,9%, erythromycin at 74%, imipenem at 41%, and ciprofloxacin resistance was found to be present in 33% of E. faecium strains (41).
In this study, 32.5% of the examined E. faecium strains were resistant to penicillin, 27.5% to erythromycin, 15% to gentamicin, 17.5% to tetracycline, 7.5% to rifampicin, 27.5% to imipenem, 5% to vancomycin, 20% to ciprofloxacin, 37.5% to clindamycin, and 17.5% to SXT. As observed in the studies, enterococci have gradually increased resistance to beta-lactam antibiotics as a result of unconscious and long-term irregular use. Besides, E. faecium strains were found to have a low resistance to tetracycline, while E. faecalis strains were found to be highly resistant. On the other hand, the rates of resistance to imipenem, ciprofloxacin, gentamicin, clindamycin, and trimethoprim/sulfamethoxazole were lower than the rates in isolates from other regions. The high efficacy of these agents shows that it is still possible to use them in this region.
5.1. Conclusions
Based on this study, the isolation rates of E. faecalis and E. faecium strains were found to be lower in Van located in eastern Turkey than in other regions. Due to the phenotypic changes in Enterococcus species, the identification reliability of epidemiological studies by PCR-based techniques was found to be relatively high. The biochemical identification of enterococci did not give reliable results, and the rate of false negativity was significant. Because of the diversity of field strains, knowing the biotype profiles could contribute significantly to the reduction of the false identification rate. Resistance rates of E. faecium and E. faecalis to antimicrobial agents were updated in the region.
References
-
1.
Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. 2014;12(10):1221-36. [PubMed ID: 25199988]. [PubMed Central ID: PMC4433168]. https://doi.org/10.1586/14787210.2014.956092.
-
2.
Pillay S, Zishiri OT, Adeleke MA. Prevalence of virulence genes in Enterococcus species isolated from companion animals and livestock. Onderstepoort J Vet Res. 2018;85(1):e1-8. [PubMed ID: 30035595]. [PubMed Central ID: PMC6238777]. https://doi.org/10.4102/ojvr.v85i1.1583.
-
3.
Kim DH, Chung YS, Park YK, Yang SJ, Lim SK, Park YH, et al. Antimicrobial resistance and virulence profiles of Enterococcus spp. isolated from horses in korea. Comp Immunol Microbiol Infect Dis. 2016;48:6-13. [PubMed ID: 27638114]. https://doi.org/10.1016/j.cimid.2016.07.001.
-
4.
Sukmawinata E, Sato W, Uemura R, Sueyoshi M. antimicrobial resistant enterococcus faecium , enterococcus faecalis , and other enterococcus species isolated from foal feces in Japan. J Equine Vet Sci. 2018;63:51-4. https://doi.org/10.1016/j.jevs.2018.01.005.
-
5.
Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence. 2012;3(5):421-33. [PubMed ID: 23076243]. [PubMed Central ID: PMC3485979]. https://doi.org/10.4161/viru.21282.
-
6.
Maasjost J, Muhldorfer K, Cortez de Jackel S, Hafez HM. Antimicrobial susceptibility patterns of enterococcus faecalis and enterococcus faecium isolated from poultry flocks in germany. Avian Dis. 2015;59(1):143-8. [PubMed ID: 26292548]. https://doi.org/10.1637/10928-090314-regr.
-
7.
Ali SA, Hasan KA, Bin Asif H, Abbasi A. Environmental enterococci: I. Prevalence of virulence, antibiotic resistance and species distribution in poultry and its related environment in Karachi, Pakistan. Lett Appl Microbiol. 2014;58(5):423-32. https://doi.org/10.1111/lam.12208.
-
8.
Skowron K, Jelenska A, Paluszak Z, Szala B. Prevalence and distribution of VRE (vancomycin resistant enterococci) and VSE (vancomycin susceptible enterococci) strains in the breeding environment. Ann Agric Environ Med. 2016;23(2):231-6. [PubMed ID: 27294624]. https://doi.org/10.5604/12321966.1203882.
-
9.
Nishiyama M, Ogura Y, Hayashi T, Suzuki Y. Antibiotic resistance profiling and genotyping of vancomycin-resistant Enterococci collected from an urban river basin in the provincial city of Miyazaki, Japan. Water. 2017;9(2):79. https://doi.org/10.3390/w9020079.
-
10.
Monticelli J, Knezevich A, Luzzati R, Di Bella S. Clinical management of non-faecium non-faecalis vancomycin-resistant enterococci infection. Focus on Enterococcus gallinarum and Enterococcus casseliflavus/flavescens. J Infect Chemother. 2018;24(4):237-46. [PubMed ID: 29396199]. https://doi.org/10.1016/j.jiac.2018.01.001.
-
11.
Kataoka Y, Umino Y, Ochi H, Harada K, Sawada T. Antimicrobial susceptibility of enterococcal species isolated from antibiotic-treated dogs and cats. J Vet Med Sci. 2014;76(10):1399-402. [PubMed ID: 24976587]. [PubMed Central ID: PMC4221175]. https://doi.org/10.1292/jvms.13-0576.
-
12.
Yılmaz EŞ, Aslantaş Ö, Önen SP, Türkyılmaz S, Kürekci C. Prevalence, antimicrobial resistance and virulence traits in enterococci from food of animal origin in Turkey. LWT Food Sci Technol. 2016;66:20-6. https://doi.org/10.1016/j.lwt.2015.10.009.
-
13.
Chajęcka-Wierzchowska W, Zadernowska A, Łaniewska-Trokenheim Ł. Virulence factors of Enterococcus spp. presented in food. Lwt. 2017;75:670-6. https://doi.org/10.1016/j.lwt.2016.10.026.
-
14.
Pinholt M, Ostergaard C, Arpi M, Bruun NE, Schonheyder HC, Gradel KO, et al. Incidence, clinical characteristics and 30-day mortality of enterococcal bacteraemia in Denmark 2006-2009: a population-based cohort study. Clin Microbiol Infect. 2014;20(2):145-51. [PubMed ID: 23647880]. https://doi.org/10.1111/1469-0691.12236.
-
15.
Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Mazumdar V, et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio. 2013;4(4). [PubMed ID: 23963180]. [PubMed Central ID: PMC3747589]. https://doi.org/10.1128/mBio.00534-13.
-
16.
Winn W, Stephan A, Janda W, Koneman EW, Procop G. Koneman's color atlas and textbook of diagnostic Microbiology. 6th ed. Baltimore, Philadelphia: Lippincott Williams & Wilkins; 2006. p. 640-3.
-
17.
Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol. 2004;42(8):3558-65. [PubMed ID: 15297497]. [PubMed Central ID: PMC497640]. https://doi.org/10.1128/JCM.42.8.3558-3565.2004.
-
18.
Kariyama R, Mitsuhata R, Chow JW, Clewell DB, Kumon H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J Clin Microbiol. 2000;38(8):3092-5. [PubMed ID: 10921985]. [PubMed Central ID: PMC87194]. https://doi.org/10.1128/JCM.38.8.3092-3095.2000.
-
19.
Carter GR. Diagnostic procedures in veterinary bacteriology and mycology. 4th ed. Springfield, Illinois, USA: C. C. Thomas; 1984.
-
20.
Lenette EH, Balows A, Hausler JWJ, Shadomy JH. Manual of clinical microbiology. 4 ed. Washington D. C., USA: American Society for Microbiology; 1985. 1149 p.
-
21.
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493-6. [PubMed ID: 5325707].
-
22.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility-23th informational supplement (CLSI Document M100-S23). Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2013.
-
23.
Nasaj M, Mousavi SM, Hosseini SM, Arabestani MR. Prevalence of virulence factors and vancomycin-resistant genes among Enterococcus faecalis and E. faecium isolated from clinical specimens. Iran J Public Health. 2016;45(6):806-13. [PubMed ID: 27648425]. [PubMed Central ID: PMC5026837].
-
24.
Mihajlović-Ukropina M, Medić D, Jelesić Z, Gusman V, Milosavljević B, Radosavljević B. Prevalence of different enterococcal species isolated from blood and their susceptibility to antimicrobial drugs in Vojvodina, Serbia, 2011-2013. Afr J Microbiol Res. 2014;8(8):819-24. https://doi.org/10.5897/ajmr2013.6517.
-
25.
Manero A, Blanch AR. Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol. 1999;65(10):4425-30. [PubMed ID: 10508070]. [PubMed Central ID: PMC91588]. https://doi.org/10.1128/AEM.65.10.4425-4430.1999.
-
26.
Facklam RR, Sahm DF. Enterococcus. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. 6th ed. Washington: ASM Press; 1995. p. 308-14.
-
27.
Pelicioli Riboldi G, Preusser de Mattos E, Guedes Frazzon AP, Alves d'Azevedo P, Frazzon J. Phenotypic and genotypic heterogeneity of Enterococcus species isolated from food in Southern Brazil. J Basic Microbiol. 2008;48(1):31-7. [PubMed ID: 18247393]. https://doi.org/10.1002/jobm.200700226.
-
28.
Sood S, Malhotra M, Das BK, Kapil A. Enterococcal infections & antimicrobial resistance. Indian J Med Res. 2008;128(2):111-21. [PubMed ID: 19001673].
-
29.
Billstrom H, Lund B, Sullivan A, Nord CE. Virulence and antimicrobial resistance in clinical Enterococcus faecium. Int J Antimicrob Agents. 2008;32(5):374-7. [PubMed ID: 18715765]. https://doi.org/10.1016/j.ijantimicag.2008.04.026.
-
30.
Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology (Reading). 2009;155(Pt 6):1749-57. [PubMed ID: 19383684]. https://doi.org/10.1099/mic.0.026385-0.
-
31.
Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. 2005;59(3):79-87. [PubMed ID: 16049560]. https://doi.org/10.18926/AMO/31979.
-
32.
Vergis EN, Shankar N, Chow JW, Hayden MK, Snydman DR, Zervos MJ, et al. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin Infect Dis. 2002;35(5):570-5. [PubMed ID: 12173131]. https://doi.org/10.1086/341977.
-
33.
Ulusoy S. Resistant gram-positive bacterial infections. Hast Inf Derg. 1999;3:212-21.
-
34.
Berzeg D, Kart Yaşar K, Şengöz G, Batı Kutlu S, Nazlıcan Ö. Antibiotic susceptibilities of enterococcus strains isolated from clinical samples. Türk Mikrobiyol Cem Der. 2005;35:279-83.
-
35.
Aktepe OC, Aşık G, Çiftçi İH, Çetinkaya Z. Antibiotic resistance rates in enterococcus strains isolated from clinical specimens. Türk Mikrobiyol Cem Derg. 2011;41(2):86-90.
-
36.
Aral M, Pakoz NIE, Aral I, Dogan S. Antibiotic resistance of enterococcus faecalis and enterococcus faecium strains isolated from various clinical samples. Türk Hij Der Biyol Derg. 2011;68(2). https://doi.org/10.5505/TurkHijyen.2011.53315.
-
37.
İraz M, Ceylan A, Akkoyunlu Y. Antibiotic susceptibility of Enterococcus strains isolated from various clinical samples. ANKEM Derg. 2012;26(4):176-80. https://doi.org/10.5222/ankem.2012.176.
-
38.
Altun D, Erdem G, Çöplü N, Çağatay M. Investigation of enterococcus strains isolated from clinical materials to species level and determination of antibiotic susceptibilities by various methods. ANKEM Derg. 2013;27(3):130-4.
-
39.
Güçkan R, Elmas A, Tilgel S, Yüksel G. Antimicrobial resistance rates of enterococcus strains isolated from various clinical specimens. Int J Basic Clin Med. 2013;1(2):74-7.
-
40.
Barišic Z, Punda-Polic V. Antibiotic resistance among enterococcal strains isolated from clinical specimens. Int J Antimicrob Agents. 2000;16(1):65-8. https://doi.org/10.1016/s0924-8579(00)00197-7.
-
41.
Schouten MA, Voss A, Hoogkamp-Korstanje JA. Antimicrobial susceptibility patterns of enterococci causing infections in Europe. The European VRE Study Group. Antimicrob Agents Chemother. 1999;43(10):2542-6. [PubMed ID: 10508041]. [PubMed Central ID: PMC89517]. https://doi.org/10.1128/AAC.43.10.2542.