Abstract
Background:
In the dairy industry, protection against species substitution or admixture is important for several reasons, including frequent human adverse reactions toward some species milk proteins, and trade and government regulations.Objectives:
The objective of the present study was to assess the purity of buffalo milk and it’s products offered as “pure buffalo” in the market.Methods:
Using species-specific primers, a duplex polymerase chain reaction (PCR) assay was performed to detect the fraudulent addition of cow’s milk to buffalo’s milk and it’s products.Results:
The limit of detection of cow’s milk in buffalo’s milk, yogurt and cheese is 1, 2 and 4%, respectively. Undeclared presence of cow’s milk was detected in 70% of the milk samples, 64% of the yogurt and 52% of the cheese samples. In 10% of the yogurt samples and 14% of the cheese samples, no apparent buffalo-related amplification product was observed, suggesting that cow’s milk was entirely substituted for buffalo’s milk in these samples.Conclusions:
To avoid unfair competition and to assure consumers of accurate labeling, using this method is recommended for regulatory agencies.Keywords
1. Background
Species identification in milk and dairy products has received special attention in the recent years. In many countries, laws require producers to clarify the type of milk used for manufacturing dairy products. Species identification of milk and dairy products is important for several reasons related to public health, religion, trade and government regulations. However, a widespread adulterant practice found in the dairy industry is the use of a less costly type of milk instead of more expensive ones. An eminent example is the addition of cow’s milk to sheep’s, goat’s or buffalo’s milk or other dairy products that are faultily labeled “pure sheep,” “pure goat,” or “pure buffalo” (1-4).
To assure consumers of accurate labeling, it is necessary to prove the authenticity of labels, using fast, reliable and sensitive methods for species identification. Among many different analytical approaches, which have been used for species identification of milk and dairy products, polymerase chain reaction (PCR)-based methods are the most reliable and sensitive techniques. Polymerase chain reaction-based methods are currently used for milk species identification, including the development of conserved mitochondrial or nuclear DNA primers for PCR amplification, followed by complementary techniques such as sequencing or PCR-Restriction fragment length polymorphism (RFLP). Alternatively, specific primers have been successfully applied for the direct detection of target species in simplex or multiplex PCR formats (1, 4-8).
In South-western Iran, water buffalo is a domestic dairy animal and buffalo’s milk and it’s products, such as yogurt and cheese, have attained great popularity. Increased demand for these products as well as higher prices, have led to the substitution or admixture of buffalo’s milk with cow’s milk.
2. Objectives
The aim of the present study was to assess the purity of buffalo products offered as “pure buffalo” in the market, using species-specific primers.
3. Methods
3.1. Samples
During a four-month period, a total of 150 samples of buffalo’s milk, yogurt and cheese were purchased from supermarkets of Ahvaz, Iran. According to the information provided by the vendors, all samples contained pure buffalo’s milk. All samples were placed in cold portable insulated boxes and transported to the laboratory.
3.2. DNA Extraction
To extract DNA from milk samples, 15 mL of milk was centrifuged at 5000 rpm for 15 minutes at room temperature. Supernatant, including the hardened fat layer and the aqueous middle phase were discarded and the bottom 1 mL of the remaining supernatant, which included somatic cells and the casein pellet was transferred to a 1.5-mL microcentrifuge tube. To dissolve casein, 300 µL of EDTA (0.5 M, pH 8) and 200 µL TE buffer (10 mM Tris-HCl, and 1 mM EDTA, pH 7.6) were added to the 1-mL supernatant. Somatic cells and casein micelles were resuspended by mixing for 30 minutes. Tubes were then centrifuged at 12000 rpm for 10 minutes at room temperature and the supernatant was discarded. Afterwards, 245 µL of lysis buffer (100 mM Tris-HCl pH 8, 200 mM NaCl, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, 5 mM ethylenediaminetetraacetic acid (EDTA)) and 5 µL of proteinase K (20 mg/mL) were added to the pellet and incubated overnight at 55°C. Subsequently the lysate was used for the usual stepwise method of DNA extraction with phenol: chloroform: isoamyl alcohol (25:24:1), and finally precipitated with one-tenth volume of sodium acetate (3 M, pH 5.2) and 2.5 volume of chilled absolute ethanol. The precipitated DNA was washed with 80% alcohol, dried and dissolved in 50 µL of sterile distilled water. The extracted DNA was frozen until PCR analysis (9, 10).
To extract DNA from yogurt samples, 18 mL EDTA (0.5 M, pH 8) and 12 mL TE buffer (10 mM Tris-HCl, and 1 mM EDTA, pH 7.6) were transferred to a 50 mL falcon tube containing 10 g of yogurt sample. After mixing for 15 minutes, samples were centrifuged at 4000 rpm for 20 minutes at room temperature. Supernatant, including the hardened fat layer and the aqueous middle phase were discarded and 1 mL of lysis buffer (100 mM Tris-HCl pH 8, 200 mM NaCl, 0.1% SDS, 1% Triton X-100, 5 mM EDTA) and 15 µl proteinase K (20 mg/ml) was added to the remaining pellet. After overnight incubation at 55°C, the lysate was used for the usual stepwise method of DNA extraction with phenol: chloroform: isoamyl alcohol (25:24:1), and ethanol precipitation as mentioned for the milk samples. The extracted DNA was frozen until PCR analysis (6, 9, 10).
To extract DNA from cheese samples, 10 mL of lysis buffer (100 mM Tris-HCl pH 8, 200 mM NaCl, 0.1% SDS, 1% Triton X-100, 5 mM EDTA) and 50 µL proteinase K (20 mg/ml) were transferred to a 50-mL falcon tube containing 5 g of cheese sample and incubated overnight at 55°C. Subsequently, the lysate was used for the usual stepwise method of DNA extraction with phenol: chloroform: isoamyl alcohol (25:24:1), and ethanol precipitation as mentioned for the milk samples. The extracted DNA was frozen until PCR analysis (6, 9, 10).
3.3. Duplex PCR Assay
Two sets of primers were used for duplex PCR according to Bottero et al., 2002. Oligonucleotide primers were 5’-GGCTTATATTACGGGTCTTACACT-3’ (forward) and 5’-GGCAATTGCTATGATGATAAATGGA-3’ (reverse) for the cow and 5’-GGCATATACTACGGATCATATACC-3’ (forward) and 5’-AATTCATTCAACCAGACTTG TACCA-3’ (reverse) for the buffalo. Amplification condition for duplex PCR was as follows: three minutes at 94°C; 35 cycles of 45 seconds at 94°C, 45 seconds at 60°C, and one minute at 72°C; and a final five-minute extension at 72°C. The presence of the 279 bp cow-specific and 192 bp buffalo-specific amplification products were checked on 1.5% agarose gel (1).
3.4. Determination of the Detection Limit
A reconstruction experiment was conducted to determine the detection limit of the assay. Samples of buffalo’s milk, yogurt and cheese containing 0.5, 1, 2, 4, 6, 8 and 10% (vol/vol) of cow’s milk were prepared. These mixed samples were then subjected to DNA extraction and subsequent duplex PCR. Finally, the detection limit of the method was estimated by agarose gel electrophoresis of the PCR products.
4. Results
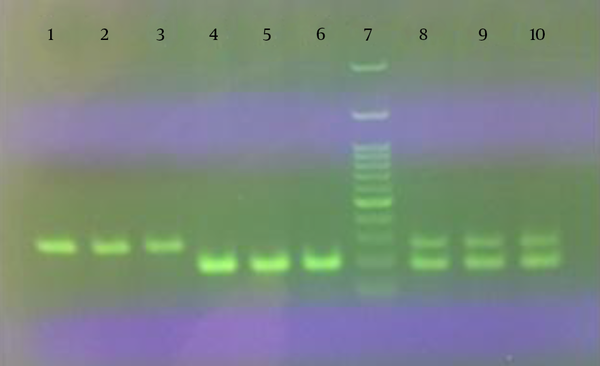
In the present study, a duplex PCR assay was used to detect the fraudulent addition of cow’s milk to the buffalo products. The extraction method used to isolate DNA from milk, yogurt and cheese samples showed good DNA yield and quality. As shown in Figure 1, simplex and duplex PCR assays on extracted DNA from binary mixture samples of milk, yogurt and cheese, resulted in 279 and 192 bp amplification products for cow and buffalo, respectively.
Results of Simplex and Duplex Polymerase Chain Reaction on Extracted DNA From Binary Mixture Samples of Milk (lanes 1, 4 and 8), Yogurt (lanes 2, 5 and 9) and Cheese (lanes 3, 6 and 10)
Results of the reconstruction experiments to determine the detection limit of the method are presented in Figure 2.
The Limit of Detection of Cow’s Milk in Buffalo’s Milk (A), Yogurt (B) and Cheese (C)
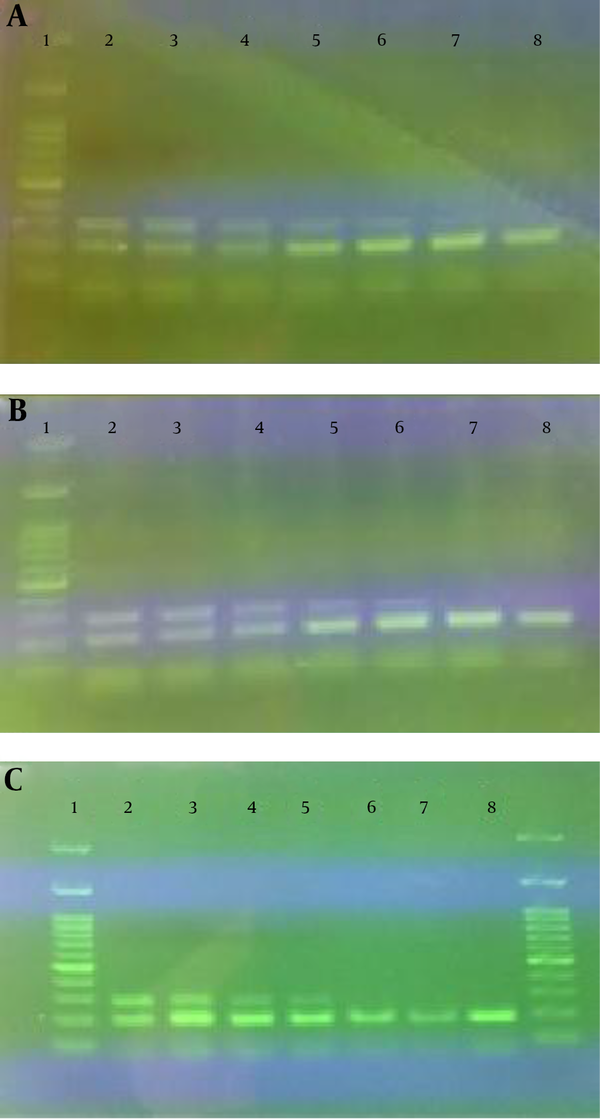
As indicated, the limit of detection of cow’s milk in buffalo’s milk, yogurt and cheese were 1, 2 and 4%, respectively.
Table 1 shows the results of PCR analyses of 150 samples of buffalo’s milk, yogurt and cheese from retail traders.
Results of Polymerase Chain Reaction Analyses of Labeled Buffalo’s Milk, Yogurt and Cheese Samples
| Sample | Total No | Pure Buffalo | Pure Cow | Mixed |
|---|---|---|---|---|
| Milk | 50 | 15 (30 %) | - | 35 (70 %) |
| Yogurt | 50 | 13 (26 %) | 5 (10 %) | 32 (64 %) |
| Cheese | 50 | 17 (34 %) | 7 (14 %) | 26 (52 %) |
Undeclared presence of cow’s milk was detected in 70% of the milk samples, 64% of the yogurt and 52% of the cheese samples. In 10% of the yogurt samples and 14% of the cheese samples, no apparent buffalo-related amplification product was observed, suggesting that cow’s milk was entirely substituted for buffalo’s milk in these samples.
5. Discussion
Illegal adulteration of raw materials used for the commercial preparation of food is a common problem. The “Farm to Fork” concept implies the traceability and authenticity of a product from raw material to consumption. To guarantee the food authenticity, the development of analytical techniques to enable authorities and producers to check if the products are correctly described and labeled is necessary. The polymerase chain reaction is the most widely used molecular technique for the identification of the species origin in food, especially in meat products (11-15). In contrast, application of PCR-based techniques for the authentication of dairy products has been very limited.
The limits of detection of the duplex PCR used in this study were 1, 2 and 4% for milk, yogurt and cheese, respectively. As non-authentic dairy products are produced for financial gain, adulterating a more expensive type of milk with a less costly type for less than 5% lacks any economic effect (16, 17). Therefore, the determined detection limits of duplex PCR assays described here are sufficient for the proof of undeclared components of the products.
Undeclared presence of cow’s milk was detected in 70% of the milk samples, 64% of the yogurt and 52% of the cheese samples. Khanzadi et al. (16) reported that only 21 out of 105 (20%) samples of sheep’s milk contained pure sheep’s milk and undeclared presence of cow’s and goat’s milk was detected in 33 (31.5 %) and 68 (65 %) of the samples, respectively. In Romania, the presence of undeclared cow’s milk was detected in 67.3% of goat and sheep cheeses (18). Maskova and Paulickova (19) found undeclared presence of cow’s milk in 3/17 of the goat cheeses and 1/7 of the sheep cheese. Di-Pinto et al. (3) analyzed 30 mozzarella cheese samples and the presence of cow’s milk was found in 22 samples. Santos et al. (20) analyzed 13 cow, goat, and sheep cheeses declared as pure. In four samples they detected the presence of an undeclared constituent, i.e. either cow’s milk or, in two cases, goat’s milk in “pure sheep cheese”. Mafra et al. (7) found that only eight out of ten ovine cheeses purchased and analyzed contained the species ingredients as listed on the package.
In conclusion, results of the present study revealed a high level of adulteration in the retail trade of buffalo products. Hence, careful and continual surveillance of the production and sale of these products should be considered. In order to achieve detailed monitoring, it is necessary to have to a fast and accurate diagnostic technique. The PCR method used in this study was a useful and straightforward approach for the detection of low levels of cows’ milk in buffalo’s milk and products. Therefore, to avoid unfair competition and to assure consumers of accurate labeling, using this method can be recommended for regulatory agencies.
Acknowledgements
References
-
1.
Bottero M, Civera T, Nucera D, Rosati S, Sacchi P, Turi R. A multiplex polymerase chain reaction for the identification of cows', goats' and sheep's milk in dairy products. Int Dairy J. 2003;13(4):277-82. https://doi.org/10.1016/S0958-6946(02)00170-X.
-
2.
Calvo J, Osta R, Zaragoza I, Zaragoza P. Species-specific amplification for detection of bovine, ovine and caprine cheese. Milchwissenschaft. 2002;57(8):444-6.
-
3.
Di Pinto A, Conversano MC, Forte VT, Novello L, Tantillo G. Detection of cow milk in buffalo "mozzarella" by polymerase chain reaction (PCR) assay. J Food Quality. 2004;27(6):428-35. https://doi.org/10.1111/j.1745-4557.2004.00662.x.
-
4.
Lopez-Calleja I, Gonzalez I, Fajardo V, Rodriguez MA, Hernandez PE, Garcia T, et al. Rapid detection of cows' milk in sheeps' and goats' milk by a species-specific polymerase chain reaction technique. J Dairy Sci. 2004;87(9):2839-45. [PubMed ID: 15375042]. https://doi.org/10.3168/jds.S0022-0302(04)73412-8.
-
5.
Abdel-Rahman S, Ahmed M. Rapid and sensitive identification of buffalo's, cattle's and sheep's milk using species-specific PCR and PCR-RFLP techniques. Food Control. 2007;18(10):1246-9. https://doi.org/10.1016/j.foodcont.2006.08.003.
-
6.
Branciari R, Nijman IJ, Plas ME, Di Antonio E, Lenstra JA. Species origin of milk in Italian mozzarella and Greek feta cheese. J Food Prot. 2000;63(3):408-11. [PubMed ID: 10716574].
-
7.
Mafra I, Ferreira IM, Faria MA, Oliveira BP. A novel approach to the quantification of bovine milk in ovine cheeses using a duplex polymerase chain reaction method. J Agric Food Chem. 2004;52(16):4943-7. [PubMed ID: 15291455]. https://doi.org/10.1021/jf049635y.
-
8.
Rea S, Chikuni K, Branciari R, Sangamayya RS, Ranucci D, Avellini P. Use of duplex polymerase chain reaction (duplex-PCR) technique to identify bovine and water buffalo milk used in making mozzarella cheese. J Dairy Res. 2001;68(4):689-98. [PubMed ID: 11928964].
-
9.
Murphy MA, Shariflou MR, Moran C. High quality genomic DNA extraction from large milk samples. J Dairy Res. 2002;69(4):645-9. [PubMed ID: 12463700].
-
10.
Sambrook J, Russell David W. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 1989.
-
11.
Ghovvati S, Nassiri MR, Mirhoseini S, Moussavi AH, Javadmanesh A. Fraud identification in industrial meat products by multiplex PCR assay. Food Control. 2009;20(8):696-9. https://doi.org/10.1016/j.foodcont.2008.09.002.
-
12.
Haunshi S, Basumatary R, Girish PS, Doley S, Bardoloi RK, Kumar A. Identification of chicken, duck, pigeon and pig meat by species-specific markers of mitochondrial origin. Meat Sci. 2009;83(3):454-9. [PubMed ID: 20416682]. https://doi.org/10.1016/j.meatsci.2009.06.026.
-
13.
Kesmen Z, Sahin F, Yetim H. PCR assay for the identification of animal species in cooked sausages. Meat Sci. 2007;77(4):649-53. [PubMed ID: 22061954]. https://doi.org/10.1016/j.meatsci.2007.05.018.
-
14.
Rodriguez MA, Garcia T, Gonzalez I, Asensio L, Hernandez PE, Martin R. PCR identification of beef, sheep, goat, and pork in raw and heat-treated meat mixtures. J Food Prot. 2004;67(1):172-7. [PubMed ID: 14717369].
-
15.
Rodriguez MA, Garcia T, Gonzalez I, Asensio L, Mayoral B, Lopez-Calleja I, et al. Identification of goose, mule duck, chicken, turkey, and swine in foie gras by species-specific polymerase chain reaction. J Agric Food Chem. 2003;51(6):1524-9. [PubMed ID: 12617577]. https://doi.org/10.1021/jf025784+.
-
16.
Khanzadi S, Jamshidi A, Razmyar J, Mohsenzadeh M. PCR-based detection of cow and goat milk in sheep milk and dairy products marketed in Mashhad city of Iran. Iranian J Veter Med. 2013;7(4):257-62.
-
17.
Maudet C, Taberlet P. Detection of cows' milk in goats' cheeses inferred from mitochondrial DNA polymorphism. J Dairy Res. 2001;68(2):229-35. [PubMed ID: 11504387].
-
18.
Stanciuc N, Rapeanu G. Identification of adulterated sheep and goat cheeses marketed in Romania by immunocromatographic assay. Food Agriculture Immunol. 2010;21(2):157-64. https://doi.org/10.1080/09540100903508683.
-
19.
Maskova E, Paulickova I. PCR-based detection of cow's milk in goat and sheep cheeses marketed in the Czech Republic. Czech Journal of Food Science Vol. 2006;24(3):127-32.
-
20.
Santos J, Fernandes P, Bardsley R. Portuguese "PDO "cheese and species origin of milk. Electro J Environmet. 2003;2:476-9.