Abstract
Background:
Escherichia coli O157:H7 (EHEC) has been traditionally associated with foodborne infections from consumption of foods with animal origin such as ground beef and burgers.Objectives:
The objective of this study was to investigate the contamination of fresh ground beef with non-sorbitol fermenting Escherichia coli and presence of virulence genes in isolates obtained from butchers located in Ahvaz, Iran.Materials and Methods:
A total of 200 fresh ground beef were sampled during a six-month period. All samples were enriched in Tryptic Soy Broth (TSB) with novobiocin and plating on Cefixime Telluride-Sorbitol MacConkey (CT-SMAC). The suspected colonies were subjected to Polymerase Chain Reaction (PCR) analysis to identify virulence genes containing rfbE O157, flic H7, stx1 and stx2 genes.Results:
Overall, 1.5% of ground beef samples were contaminated with the O157 E. coli strain meanwhile 1% of samples contained the O157:H7 strain and 0.5% of samples had the O157:H7 with virulent stx1 and stx2 genes.Conclusions:
The observed results indicated the necessity of good care in abattoir, butcheries and during food distribution, mainly ground beef. Also, a zero tolerance policy could be useful to control E. coli O157 in meat products nationwide.Keywords
Polymerase Chain Reaction (PCR) Isolation Meat Products Molecular Characterization Escherichia coli
1. Background
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is one of the most important foodborne pathogens that produces verotoxin and has been traditionally associated with foodborne infection from consumption of foods with animal origin, particularly those originating from cattle, such as ground beef and burgers (1). Hemorrhagic Colitis (HC) and Hemolytic Uremic Syndrome (HUS) are two chronic and potentially fatal illnesses caused by EHEC (2). Consumption of undercooked minced beef is the most common route of transmission of EHEC. Thousands of illnesses and hospitalizations and hundreds of deaths due to this bacterium has been reported (3).
To the best of our knowledge, isolation of the pathogenic organism from ground beef and meat products in various countries such as Republic of Ireland (4), Egypt (5), Switzerland (6), Turkey (7), Mexico (8), USA (9), Brazil (10) and China (11) has been documented. In Iran, there has been a number of assays on the isolation and the prevalence of E. coli O157:H7 in ground beef and beef products. Jafareyan reported that 10 (6.8%) of the 148 ground beef samples were contaminated with E. coli O157:H (12). Shekarforoush et al. detected verotoxigenic E. coli O157:H7 in six (3.92%) of the 153 sheep carcass samples (13) and Shahrokhabadi found that six (4.05%) of the 148 bovine carcasses samples were contaminated With the bacterium (14).
The pathogenicity of E. coli O157:H7 is affected by several virulence factors. The main factor contributing to the pathogenicity is its ability to produce potential cytotoxins called Shiga-toxins (Stx), encoded by stx1 and stx2 genes (15). Other described virulence factors include intimin, encoded by the eaeA gene and EHEC hemolysin encoded by the EHEC hlyA gene (16). Also, it has been reported that non-O157 strains could be verocytotoxigenic (17, 18).
2. Objectives
The aim of the present study was to determine the prevalence of viable Non-Sorbitol Fermenting (NSF) E. coli containing rfbE O157 and flic H7 genes and, to detect the presence of the stx1 and stx2 genes in isolates from ground beef samples obtained from butcheries in Ahvaz city, Iran.
3. Materials and Methods
3.1. Sample Preparation
A total of 200 samples of fresh ground beef were obtained from butcheries located in different parts of Ahvaz during a six-month period. Samples were transported to the laboratory under cold conditions. Samples were analyzed according to E. coli O157:H7 isolation procedures on the same day.
3.2. Isolation and Identification of Non-Sorbitol Fermenting Escherichia coli
All samples were examined for the presence of NSF E. coli by regular procedures (19). Ten grams of each sample was added to 90 mL Tryptone Soy Broth (TSB) (Quelab, Canada) supplemented with novobiocin (20 mg/L, Sigma) and homogenized. After incubation for 24 hours at 37˚C, a 100-µL suspension was spread onto Cefixime Telluride-Sorbitol MacConkey (CT-SMAC) agar (scharlau, Spain) supplemented with cefixime (0.25 mg/mL) and tellurite potassium (2.5 mg/mL). Plates were incubated for 24 hours at 37˚C and examined for typical E. coli colonies (colorless, circular with brown center). The suspected NSM E. coli colonies were plated on both Eosin-Methylene Blue agar (EMB) (Scharlau, Spain) and Tryptone Bile x-Glucoronide (TBX) (Merck, Germany) and were incubated at 37°C for 24 hours. Also, the suspected isolates were subjected to Polymerase Chain Reaction (PCR) analysis to identify virulent genes.
3.3. The Polymerase Chain Reaction Procedures
3.3.1. DNA Extraction
Extraction was performed using a modification of the method previously described by Lopez-Saucedo et al. (20). Presumptive E. coli colonies were separately grown overnight in 5 mL of TSB at 37°C. These cultures were centrifuged (Hitachi 1110, Germany), the pellet was resuspended in 1 mL of sterile distilled water, and samples were heated at 100°C for ten minutes. After heating, the suspension was again centrifuged and the supernatant was used as the PCR template.
3.3.2. Polymerase Chain Reaction and Electrophoresis
Presumptive colonies in two steps were subjected to the PCR assay for amplification of four pairs of specific primers, including: (FliC) H7 and O157 in first and stx1 and stx2 in the second steps, according to the following program for both steps: initial denaturation at 94˚C for three minutes, and then 35 cycles comprised of denaturation at 94˚C for 45 seconds, annealing at 60˚C for 45 seconds, and an extension at 72˚C for 60 seconds. Following this, a final extension at 72˚C for five minutes was carried out. Each PCR tube contained 25 μL of reaction mixture, consisting of 2.5 µL of PCR buffer (10x), 1.5 µL MgCl2 (50 mM), 1 µl dNTP (10 mM), 0.5 µl Taq polymerase (2.5 U), 4 µL of a mixture of the two forward and revised primers (15 µM), 10.5 µL of ddH2O and 5 μL of template extracted DNA. The mixture was then processed in a thermocycler (Bioer Technology Co., China). The targets, primer sequences and amplicon sizes for the PCR products are shown in Table 1.
List of Target Genes, Sequence of Primers and Product Size (bp)
| Primer | Size (bp) | Sequence | Reference |
|---|---|---|---|
| O157 | 259 | (21) | |
| F: 5 –CGGACATCCATGTGATATGG-3 | |||
| R: 5-TTGCCTATGTACAGCTAATCC -3 | |||
| (FliC) H7 | 625 | (22) | |
| F : 5- GCGCTGTCGAGTTCTATCGAGC -3 | |||
| R: 5- CAACGGTGACTTTATCGCCATTCC -3 | |||
| STX1 | 614 | (23) | |
| F: 5 – ACACTGGATGATCTCAGTGG-3 | |||
| R: 5- CTGAATCCCCCTCCATTATG -3 | |||
| STX2 | 779 | (23) | |
| F : 5- CCATGACAACGGACAGCAGTT -3 | |||
| R: 5- CCTGTCAACTGAGCAGCACTTTG -3 |
The amplified PCR products were detected by electrophoresis (Paya pajoohesh, Iran) and staining, and visualized under UV light illumination (UVT-20 SL, Iran).
4. Results
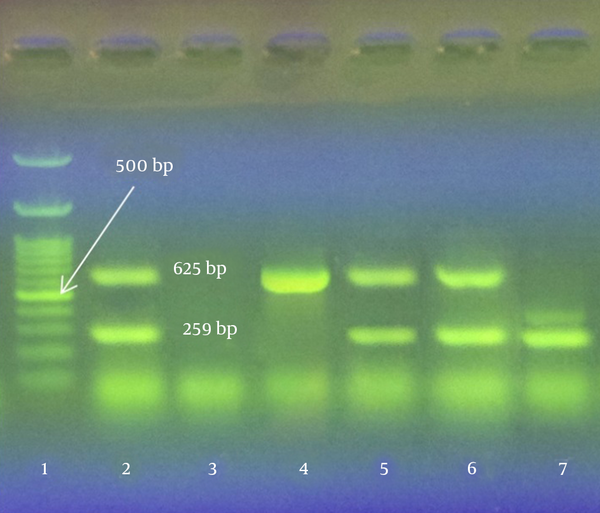
The present study focused on identification of virulent genes in NSF isolates from ground beef in Ahvaz city. A total of 23 strains of NSF E. coli were recovered from 200 meat samples (11.5%) by CT-SMAC culture agar and plating on EMB agar. Among these, 14 strains were colorless on TBX medium and probably had negative ß-glucuronidase enzyme activity. Our findings showed that two strains contained both O157 and H7 genes, one strain was O157 and nine strains were only H7 positive. Virulent stx1 and stx2 genes were found only in an O157:H7 strain (Figures 1 and 2).
Polymerase Chain Reaction Results on Gel Electrophoresis
Polymerase Chain Reaction Results for stx Detection on Gel Electrophoresis
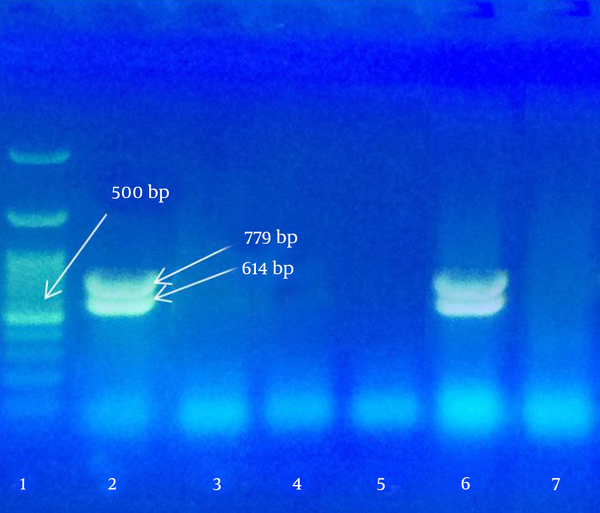
In conclusion, 1.5% of ground beef samples contaminated the O157 E. coli strain meanwhile 1% of samples were O157:H7 positive and 0.5% of samples were O157:H7 positive containing virulent stx1 and stx2 genes.
5. Discussion
Beef is one of the favorable consumed meats in the world. It has been noted that beef carcass and ground beef are the most important sources of E. coli O157:H7 (24). Infections caused by these bacteria could lead to diarrhea, hemorrhagic colitis or Hemolytic Uremic Syndrome (HUS) (25). The bacterium recognized as low dose foodborne pathogens (17).
In our study, serovar O157:H7, not motile O157 and non-O157 E. coli were isolated from two (1%), one (0.5%) and 20 (10%) ground beef samples, respectively, which indicates the risk of consumption of this kind of food if severely undercook. Several studies have shown that E. coli O157:H7 and other Shiga-Toxin E. coli (STEC) are present in meat products; mostly beef products. For example Cagney et al. (4) investigated the prevalence and numbers of E. coli O157:H7 in minced beef and beef burgers in supermarkets and butcheries in the Republic of Ireland. Overall, E. coli O157:H7 was recovered from 43 samples (2.80%). In France, Vernozy-Rozand et al. reported that 0.12% (4/3450) of samples were positive for E. coli O157:H7 in large-scale processed minced beef (26). Escherichia coli non-O157, E. coli O157: NM (not motile) and E. coli O157:H7 were isolated from 53 (20.5%), 13 (5%) and seven (2.7%) of the 258 beef carcasses, respectively, sampled by Varela-Hernandez in Mexico (8). Ahmed and Shimamoto recovered E. coli O157:H7 from 4.3% ground beef collected from butcheries in Egypt (5). Fantelli and Stephan detected O157:H7 in 2.3% of 213 ground beef samples (6), while this pathogen was isolated from 7.6% of 251 ground beef samples by Sarimehmetoglu, in Turkey (7). In Argentina, Chinen isolated E. coli O157:H7 from 3.8% of 161 ground beef samples (27). Other studies found very different results, ranging from 16.8% (50/296) E. coli O157:H7 samples in Washington State, USA (28) to 0%, as determined by the study of Tarr et al., which did not recover the pathogen from 1400 retail ground minced beef samples from six stores in Seattle, USA (29).
A few studies have indicated that the isolation of the bacterium in meat products in Iran. For example Rahimi (30) reported a high prevalence of E. coli O157:H7 in beef samples (8.2%), followed by water buffalo (5.3%), sheep (4.8%), camel (2.0%) and goat (1.7%). Again, in another study from Iran, high incidence of E. coli O157:H7 in ruminant's meat samples was reported by Momtaz, where, 238 (29.02%) samples were positive for the presence of E. coli. All of the isolates had more than one virulence gene including stx1, stx2, eaeA and hly (31). In South-West of Iran, E. coli O157:H7 was found in six (3.92%) of 153 sheep carcasses (13). In Isfahan, Jafareyan-Sedigh reported that 10 (6.8%) of 148 sheep meat samples were contaminated with E. coli O157:H7 (12). Shahrokhabadi recovered the pathogen from six (4.05%) of 148 cattle carcasses in Slaughterhouse of Rafsanjan (14).
Various factors such as verocytotoxin (encoded by stx1 and stx2), a protein called intimin (encoded by eaeA gene) and enterohemolysin (encoded by EHEC hlyA gene) are linked to the pathogenesis of E. coli O175:H7 (16, 21, 31). In the current study, 20 non-O175 E. coli with no virulent stx1 and stx2 genes were isolated. Meanwhile, it has been reported that non-O157 strains could also be verocytotoxigenic (17, 18). Our data showed that 14 strains were colorless on TBX medium and were assumed to be the O157 strain yet only three of them contained O157 genes. It was concluded that this medium could not be a reliable medium for the detection of O157 strains. Also, the H7 gene was identified in nine non-O157 strains and may belong to the O55 strain because the flic genes of O55:H7 and O157:H7 strains are closely related (32).
Generally, 1.5% of ground beef samples were contaminated with O157 E.coli. These kinds of products may pose risks to the health of consumers if eaten raw or undercooked. To reduce risk, the incorporation of all agents involved in the beef supply chain is necessary. Also, application of Hazard Analysis of Critical Control Points (HACCP) from the farm to the abattoir should be applied by governmental authorities. Consumers and retailers should be educated about the disease and methods of prevention. A zero tolerance policy could be useful to control E. coli O157 in meat products in the country.
Acknowledgements
References
-
1.
Williams RC, Isaacs S, Decou ML, Richardson EA, Buffett MC, Slinger RW, et al. Illness outbreak associated with Escherichia coli O157:H7 in Genoa salami. E. coli O157:H7 Working Group. CMAJ. 2000;162(10):1409-13. [PubMed ID: 10834043].
-
2.
Jay JM. Food Microbiology. New York: an Nostrand reinhold; 1992. https://doi.org/10.1007/978-94-011-6480-1.
-
3.
Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5(5):607-25. [PubMed ID: 10511517]. https://doi.org/10.3201/eid0505.990502.
-
4.
Cagney C, Crowley H, Duffy G, Sheridan JJ, O’Brien S, Carney E, et al. Prevalence and numbers of Escherichia coli O157: H7 in minced beef and beef burgers from butcher shops and supermarkets in the Republic of Ireland. Food Microbiol. 2004;21(2):203-12.
-
5.
Ahmed AM, Shimamoto T. Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157:H7 and Shigella spp. from meat and dairy products in Egypt. Int J Food Microbiol. 2014;168-169:57-62. [PubMed ID: 24239976]. https://doi.org/10.1016/j.ijfoodmicro.2013.10.014.
-
6.
Fantelli K, Stephan R. Prevalence and characteristics of shigatoxin-producing Escherichia coli and Listeria monocytogenes strains isolated from minced meat in Switzerland. Int J Food Microbiol. 2001;70(1-2):63-9. [PubMed ID: 11759763].
-
7.
Sarimehmetoglu B, Aksoy MH, Ayaz ND, Ayaz Y, Kuplulu O, Kaplan YZ. Detection of Escherichia coli O157: H7 in ground beef using immunomagnetic separation and multiplex PCR. Food Control. 2009;20(4):357-61.
-
8.
Varela-Hernandez JJ, Cabrera-Diaz E, Cardona-Lopez MA, Ibarra-Velazquez LM, Rangel-Villalobos H, Castillo A, et al. Isolation and characterization of Shiga toxin-producing Escherichia coli O157:H7 and non-O157 from beef carcasses at a slaughter plant in Mexico. Int J Food Microbiol. 2007;113(2):237-41. [PubMed ID: 17007951]. https://doi.org/10.1016/j.ijfoodmicro.2006.06.028.
-
9.
Ekiri AB, Landblom D, Doetkott D, Olet S, Shelver WL, Khaitsa ML. Isolation and characterization of shiga toxin-producing escherichia coli serogroups O26, O45, O103, O111, O113, O121, O145, and O157 shed from range and feedlot cattle from postweaning to slaughter. J Food Prot. 2014;77(7):1052-61. [PubMed ID: 24988009]. https://doi.org/10.4315/0362-028X.JFP-13-373.
-
10.
Lucatelli A, editor. First Isolation of Shiga Toxin-producing Escherichia coli O157: H7 in Ground Beef at Retail Market in Sao Paulo City, Brazil. Annual Meeting. 2012; United State. Iafp;
-
11.
Xu JG, Quan TS, Xiao DL, Fan TR, Li LM, Wang CA. Isolation and characterization ofEscherichia coli O157:H7 strains in China. Current Microbio. 1990;20(5):299-303.
-
12.
Jafareyan-Sedigh M, Rahimi E, Doosti A. Isolation of Escherichia coli O157: H7 in sheep meats using cultural and PCR method. J Shahrekord U Med Sci. 2011;13(2):61-8.
-
13.
Shekarforoush S, Tahamtan Y, Pourbakhsh A. Detection and frequency of Stx2 gene in Escherichia coli O157 and O157:H7 strains isolated from sheep carcasses in Shiraz-Iran. Pak J Biol Sci. 2008;11(8):1085-92. [PubMed ID: 18819545].
-
14.
Shahrokhabadi R, Rahimi E, Momtaz H, Poursahebi R. Identification of Escherichia coli O157: H7 on Bovine Carcasses in Rafsanjan by Using Culture and PCR Method . 2012;1(8):49-53. J Veterinary Microbiol. 2012.
-
15.
Gyles CL. Shiga toxin-producing Escherichia coli: an overview. J Anim Sci. 2007;85(13 Suppl):E45-62. [PubMed ID: 17085726]. https://doi.org/10.2527/jas.2006-508.
-
16.
Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142-201. [PubMed ID: 9457432].
-
17.
Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2(1):15-38. [PubMed ID: 2644022].
-
18.
March SB, Ratnam S. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1986;23(5):869-72. [PubMed ID: 3519658].
-
19.
Carney E, O'Brien SB, Sheridan JJ, McDowell DA, Blair IS, Duffy G. Prevalence and level of Escherichia coli O157 on beef trimmings, carcasses and boned head meat at a beef slaughter plant. Food Microbiol. 2006;23(1):52-9. [PubMed ID: 16942986]. https://doi.org/10.1016/j.fm.2004.12.001.
-
20.
Lopez-Saucedo C, Cerna JF, Villegas-Sepulveda N, Thompson R, Velazquez FR, Torres J, et al. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg Infect Dis. 2003;9(1):127-31. [PubMed ID: 12533296]. https://doi.org/10.3201/eid0901.010507.
-
21.
Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36(2):598-602. [PubMed ID: 9466788].
-
22.
Gannon VP, D'Souza S, Graham T, King RK, Rahn K, Read S. Use of the flagellar H7 gene as a target in multiplex PCR assays and improved specificity in identification of enterohemorrhagic Escherichia coli strains. J Clin Microbiol. 1997;35(3):656-62. [PubMed ID: 9041407].
-
23.
Gannon VP, King RK, Kim JY, Thomas EJ. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coli in ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58(12):3809-15. [PubMed ID: 1476425].
-
24.
Lee GY, Jang HI, Hwang IG, Rhee MS. Prevalence and classification of pathogenic Escherichia coli isolated from fresh beef, poultry, and pork in Korea. Int J Food Microbiol. 2009;134(3):196-200. [PubMed ID: 19665813]. https://doi.org/10.1016/j.ijfoodmicro.2009.06.013.
-
25.
Banatvala N, Griffin PM, Greene KD, Barrett TJ, Bibb WF, Green JH, et al. The United States National Prospective Hemolytic Uremic Syndrome Study: microbiologic, serologic, clinical, and epidemiologic findings. J Infect Dis. 2001;183(7):1063-70. [PubMed ID: 11237831]. https://doi.org/10.1086/319269.
-
26.
Vernozy-Rozand C, Ray-Gueniot S, Ragot C, Bavai C, Mazuy C, Montet MP, et al. Prevalence of Escherichia coli O157:H7 in industrial minced beef. Lett Appl Microbiol. 2002;35(1):7-11. [PubMed ID: 12081541].
-
27.
Chinen I, Tanaro JD, Miliwebsky E, Lound LH, Chillemi G, Ledri S, et al. Isolation and characterization of Escherichia coli O157:H7 from retail meats in Argentina. J Food Prot. 2001;64(9):1346-51. [PubMed ID: 11563511].
-
28.
Samadpour M, Kubler M, Buck FC, Depavia GA, Mazengia E, Stewart J, et al. Prevalence of Shiga toxin-producing Escherichia coli in ground beef and cattle feces from King County, Washington. J Food Prot. 2002;65(8):1322-5. [PubMed ID: 12182487].
-
29.
Tarr PI, Tran NT, Wilson RA. Escherichia coli O157:H7 in retail ground beef in Seattle: results of a one-year prospective study. J Food Prot. 1999;62(2):133-9. [PubMed ID: 10030631].
-
30.
Rahimi E, Kazemeini HR, Salajegheh M. Escherichia coli O157:H7/NM prevalence in raw beef, camel, sheep, goat, and water buffalo meat in Fars and Khuzestan provinces, Iran. Vet Res Forum. 2012;3(1):15-7. [PubMed ID: 25653740].
-
31.
Momtaz H, Safarpoor Dehkordi F, Rahimi E, Ezadi H, Arab R. Incidence of Shiga toxin-producing Escherichia coli serogroups in ruminant's meat. Meat Sci. 2013;95(2):381-8. [PubMed ID: 23747633]. https://doi.org/10.1016/j.meatsci.2013.04.051.
-
32.
Wang L, Rothemund D, Curd H, Reeves PR. Sequence diversity of the Escherichia coli H7 fliC genes: implication for a DNA-based typing scheme for E. coli O157:H7. J Clin Microbiol. 2000;38(5):1786-90. [PubMed ID: 10790100].