Abstract
Keywords
Pseudomonas aeruginosa Infection Antibiotic Pathogenicity Ciprofloxacin
1. Background
Pseudomonas aeruginosa is a nosocomial, aerobic, gram-negative bacterium that causes pneumonia, urinary tract infections, surgical site infections, ulcerative keratitis, and soft tissue infections, among other things (1). The species are often isolated from soil and water, or they colonize a variety of anatomical sites, including plants, insects, animals, and humans (2). Virulent factors are molecules produced by pathogenic P. aeruginosa that allow it to play a significant role in pathogenesis infection. This bacterium is normally not pathogenic, but it can cause opportunistic infections in patient with weak immune systems, such as ICU patients (3). Pseudomonas aeruginosa is caused by the production of several cellular and extracellular virulence factors. Pseudomonas aeruginosa has a higher proportion of predicted regulatory genes due to its large genome of 6.3 million base pairs which suggests that P. aeruginosa can adapt to a variety of environmental stresses (4). Some of the genes that normally encode and participate in virulence factors that may contribute to its pathogenicity are toxA, exoS, exoY, exoU, oprL, oprI, lasA, lasB, oprD, plcH, plcN, and nan1. The major outer membrane lipoproteins responsible for identifying P. aeruginosa infections are oprL, oprI, and oprD. Similarly, the toxA gene, which encodes exotoxin A and inhibits protein biosynthesis, is the most toxic virulence gene. Elastase B (lasB) is an important metalloenzyme that plays a key role in pathogenesis during host infection by promoting tissue adhesion, colonization, and invasion, resulting in chronic pulmonary inflammation (5). It causes elastin lysis, protein compound destruction, complement inactivation, and clotting factor degradation (6). Exotoxin A (exoA) is responsible for tissue necrosis (7). ExoA is the major member of the type II secretion system (T2SS) which inhibits protein synthesis through ADP-ribosylation of eukaryotic elongation factor 2 (8).
Rapid detection of such virulence genes which cause hospital infections is important for subsequent treatment decisions of patients. Previously, common methods of bacterial typing such as antibiotic resistance patterns, phage typing, and serotyping were used to study the genetic linkages of bacteria, particularly in nosocomial infections. These are now being replaced by molecular methods such as ribotyping, pulse-field gel electrophoresis (PFGE), and PCR-based methods. Molecular methods are superior over phenotypic methods for identifying P. aeruginosa by developing a multiplex Polymerase chain reaction (PCR) (9). PCR is used to determine the contribution of several virulence genes in resistant mechanisms, which aids in the selection of appropriate antibiotics for the treatment of P. aeruginosa-caused infectious disease. PCR can be rapidly identify the microbial species by the amplification of sequences unique to a specific organism. The enterobacterial repetitive intergenic consensus (ERIC)-PCR technique uses genetic marker i.e., the position and number of different ERIC sequences in bacteria, to detect the bacterial diversity (10).
Antimicrobial resistance (AMR) is a public health issue that affects both human and veterinary medicine. A diverse group of P. aeruginosa strains have the inherent ability to form biofilms. And these biofilms boosts resistance to various classes of antibiotics or antibacterial agents, making infection control difficult (11). AMR in P. aeruginosa is usually the result of a combination of different imported (mobile genetic elements) and chromosomally encoded resistance mechanisms (12). Pseudomonas aeruginosa's major antibiotic-resistance mechanisms can be divided into three categories: intrinsic, acquired, and adaptive resistance. The unreasonable administration of antibiotics in both human medicine and animal production for growth-promoting purposes, metaphylaxis, and prophylaxis have nourished the proliferation, spread of antibiotic-resistant bacteria, and resistance genes. Such an uncontrolled and random use of antibiotics increased public health and environmental risks (13). And also it is difficult to cure the patients whose infections are resistant to conventional antibiotics. Hence there is an urgent need to develop the advance antibiotics and effective alternative therapeutic strategies for the treatment of P. aeruginosa infections (14).
2. Objectives
The objective of this study was to look into the virulence factors and antibiotic susceptibility isolated from animal and human samples in order to determine the pathogenicity of P. aeruginosa using the PCR technique.
3. Methods
3.1. Sample Collection and Bacterial Isolation
The study was performed on a total of 120 specimens of P. aeruginosa strains. Among of, 60 animal specimens were collected from veterinary microbiology laboratories and the remaining 60 human specimens were collected from hospital laboratories. After that, the samples were then cultured on MacConkey media, eosin methylene blue EMB agar, and incubated overnight at 33°C. Following incubation, the bacterial isolates were subjected to biochemical tests to determine bacterial growth and colony formation. Finally, 76 isolates were identified by gram staining and biochemical tests of oxidase, catalase, motion, indole sulfide (SIM), methyl (TSI) triple sugar agar-agar, (VP) Vegs Prosquier, (MR) red, (OD) ornithine decarboxylase, (SiMc) Simon citrate agar, (AD) arginine dehydrogenase, (LD) lysine decarboxylation, oxidation -fermentation test (Merck, Germany). During the period of study, 76 isolates of Pseudomonas species were identified, out of which 24 isolates of P. aeruginosa were recovered from animal samples and 52 strains were recovered from human samples. These 52 strains recovered from human clinical infection samples includes isolates of wound, urine, blood, and sputum. Detected P. aeruginosa bacterial isolates were used for subsequent tests.
3.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was determined by disk diffusion method in Müller-Hinton agar medium examined according to Clinical and Laboratory Standard Institute (CLSI) guidelines (15). The following antibiotics disks (Kirby-Bauuer) were used for antibiotic susceptibility testing: (1) ceftazidim, (2) gentamycin, (3) pipracillin, (4) amikacin, (5) ciprofloxacin, (6) imipenem, (7) cetotaxim, (8) erythromycin, (9) efloxacin, (10) tubramycin, (11) clindamycin, and (12) tetracyclin. The isolates were divided into three groups: (1) sensitive (S), (2) intermediate (I), and (3) resistant (R).
3.3. DNA Extraction and Molecular Detection of Virulence-Related Genes Using PCR Technique
Bacterial genomic DNAs were extracted from P. aeruginosa isolates using a commercial DNA extraction kit (CinnaGen Co., Tehran, Iran). Table 1 shows two sets of primers used to amplify the genes [exoA (396bp) and lasB (300bp)] using a specific polymerase chain reaction (PCR). PCR amplification was carried out using a thermal cycler (Eppendorf, Germany) with specific forward and reverse primers with specific length coding for exoA (exoenzyme A) and lasB (elastase) virulence genes. Each PCR mixture was prepared in a final volume of 25 µL containing 2.5 µL of PCR-10x buffer, 1 µL MgCl2 (2.5 mM), 1 µL of each primer (10 pmol), 1.5 µL dNTPS (2.5 mM), 0.3 µL of Tq polymerase enzyme (5U), and 2 µL of DNA template. PCR amplifications were carried out on a thermocycler under the following conditions: 95°C for 15 minutes; followed by 35 cycles at 95°C for 1 minute, 60°C for 45 seconds, 72°C for 1 minute and a final extension step at 72°C for 7 minutes. Multiple negative (water) amplification was used as a negative control (16).
Nucleotide Sequences of Primers Used for Amplification of Pseudomonas aeruginosa Virulence
4. Results
4.1. Bacterial Isolates and Distribution of Virulence-Related Genes
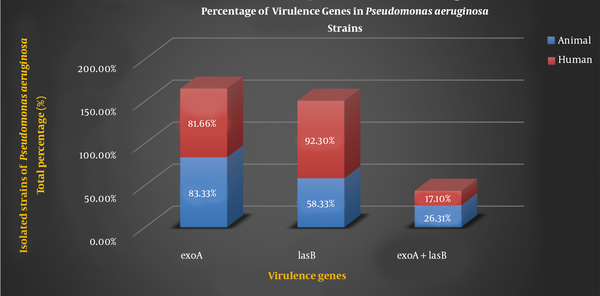
A total of 76 isolates were screened for the presence of different virulence genes of P. aeruginosa using conventional methods such as growth characteristics, colony morphology, and biochemical tests. The current study included 24 (31.57%) P. aeruginosa isolates from animal samples and 52 (68.42%) isolates from human samples. Table 2 shows the different percentages of virulence genes in P. aeruginosa strains of animal and human samples.
Percentage of Virulence Genes in Pseudomonas aeruginosa Strains
| Virulence Genes | Isolated Strains of P. aeruginosa Total Percentage (%) | |
|---|---|---|
| (Animal Sample Isolates) 24 | (Human Sample Isolates) 52 | |
| exoA | 83.33 | 81.66 |
| lasB | 58.33 | 92.30 |
| exoA + lasB | 26.31 | 17.10 |
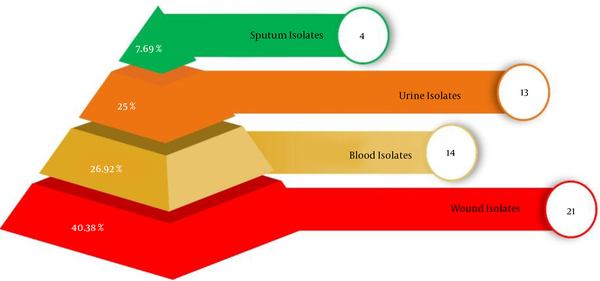
Figure 1 shows the percentage of P. aeruginosa strains isolated from a variety of clinical sites of human. Where, in human, the percentage of P. aeruginosa in blood was 26.92% (14 isolates), in the urine was 25% (13 isolates), in wound 40.38% (21 isolates), in sputum was 7.69% (4 isolates). Phenotypic analysis shows that highest percentage of P. aeruginosa strains were isolated from blood isolates.
Percentage Pseudomonas aeruginosa strains isolated from a variety of clinical sites of human.
The genes of P. aeruginosa are considered as important virulence factor attributed to the bacterium pathogenicity which are detected by PCR. The PCR results of virulence factor occurrence showed that exoA and lasB genes were present in both the animal samples and the human samples. Figure 2 shows the percentage of virulence genes in P. aeruginosa strains present in animal and human samples.
Percentage of virulence genes in Pseudomonas aeruginosa strains present in animal and human samples.
4.2 Antibiotic Resistance Patterns
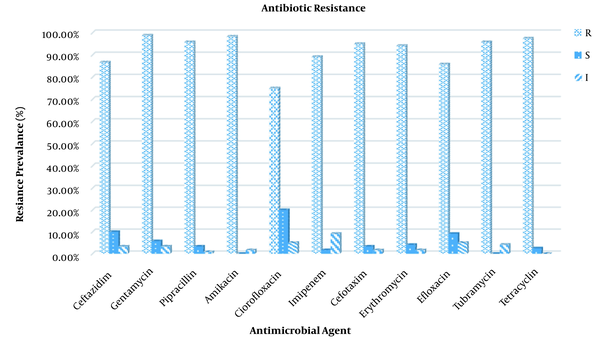
The antibiotic susceptibility testing results of P. aeruginosa strains revealed varying levels of resistance, as shown in Figure 3. According to the findings of this study, Ciprofloxacin and Tubramycin were the least resistant antibiotics against both animal and human isolates, with resistance rates of 75 and 75.53%, respectively. In contrast, all the P. aeruginosa isolates were most resistive against Amikacin (98.33%) and Tetracycline (97.50%).
The antibiotic resistance (%) of isolated Pseudomonas aeruginosa strains.
5. Discussion
In this study, the prevalence of P. aeruginosa isolates was 76, with the distribution in animal samples 24 (31.57%) was lower than in human samples 52 (68.42%). Possible explanations include different types of studied populations, different geographical locations, and different types of hospitals and veterinary laboratories. Pseudomonas aeruginosa is an opportunistic human pathogen competent for a wide array of infections including respiratory tract, blood, urinary tract, and skin infections. This competence for infections has made the organism to be recognized as a threat to public health (18). Phenotypic analysis shows that of a number of P. aeruginosa strains obtained from a variety of clinical sources, including wounds, urine, sputum, and blood. It is observe from the present study that wound patients are more liable to get infections in comparison with other patients. This was attributed to the organisms' virulence and invasive capability, the physiological state of the tissue in the wound, and the host's immunological integrity (19), lack of general hygienic measures, larger production of low quality antiseptic and medicinal solutions for treatment, difficulties in properly defining responsibilities among hospital staff (20). Wound infections are difficult to heal, and this bacterium has caused serious economic problems. Pseudomonas aeruginosa is a major cause of wound morbidity and mortality worldwide (21).
Pseudomonas aeruginosa is a pathogen having different virulence factors such as exotoxin A, alginate, exoenzyme S, elastase, and phospholipase. These virulence factors are regulated by particular signaling systems and can cause nosocomial problems to the world health system (22). Pathogenicity of P. aeruginosa is multifactorial. ExoA is responsible for the toxigenic trait of P. aeruginosa and plays a distinct role in the hindrance of wound contraction and healing. lasB, one of the most important proteases of P. aeruginosa, is responsible for invasiveness. lasB can facilitate bacterial attachment and immune system disruption (23). lasB causes elastin lysis, protein compound destruction, complement inactivation, and clotting factor degradation (6). Because of the importance of P. aeruginosa, genes encoding virulence factors including exoA, lasB genes as well as antibiotic resistance patterns were investigated. Herein, all examined isolates have exoA and lasB genes. The frequency of exoA genes in isolated strains P. aeruginosa strains was 83.33% (animal sample) and 81.66% (human sample). However, the frequency of lasB genes was found to be 58.33 and 92.30% in animal and human samples respectively. Furthermore, 26.31% of animal strains and 17.10% of human strains contain both the exoA and lasB genes (Figure 2). Divergences in the distribution of virulence factor genes across populations may be due to that some P. aeruginosa strains are better adapted to the specific conditions found in infectious sites. The prevalence of P. aeruginosa and its virulence genes is affected by a variety of factors, including the nature of the environment, the degree of contamination and type, and the individual's immune status (24). There are various reports on the frequency of exoA and lasB genes in different studies. Amirmozafar et al. detected the exoA strains in 81% of clinical isolates of P. aeruginosa (25). Neamah discovered 45.7% of exoA gene in otitis media and 75% in cow milk isolates of P. aeruginosa (26). Dekhil recorded a 54.5% prevalence of exoA gene in P. aeruginosa isolates (27) which were less than our results. According to the findings of our and other studies, exoA is a more common virulence factor. Sabharwal et al. reported the prevalence of lasB in 75% clinical isolates of P. aeruginosa (28). Comoé detected a high prevalence of elastase (89.2%) encoding the lasB genes of P. aeruginosa. This suggests that this protease, which cleaves elastin and collagen, may be important in the pathogenesis of P. aeruginosa isolated from animal products (2). Faraji et al. discovered the higher prevalence of lasB in P. aeruginosa isolated from cystic fibrosis (CF) 62 (95.4%) and burn wounds 47 (82%) than any other genes detected (22).
As P. aeruginosa is an opportunistic pathogen involved in clinical infections, it is critical to understand the full extent of the variation in antimicrobial resistance gene content (29). In the current study, 76 isolates of P. aeruginosa were tested using disc diffusion against 11 different antibiotics in order to gain a better understanding of the antibiotic resistance rates (Figure 1). Pseudomonas aeruginosa isolates showed high-level resistance to many antimicrobial agents. In the present study, the in vitro antibiotic resistivity test of isolated P. aeruginosa strains revealed the highest resistance in human and animal samples was to amikacin and there was no significant difference between them. On the other side, the lowest resistance was to ciprofloxacin and there was a significant difference between human and animal strains. Antibiotic resistance in P. aeruginosa could be caused by indiscriminate antibiotic use, the production of various enzymes such as carbapenamase and AmpC-lactamases, quorum sensing modification of different target sides, and so on (30, 31). A cause for concern is that the high resistance displayed to the ciprofloxacin which is one of the best options available for the treating the infections caused by P. aeruginosa, particularly in the treatment of urinary tract infections. Such a drwabacks will result in limited treatment options (32). In 2016, Sharma et al. (33) discovered that amikacin (18.2% ) and ciprofloxacin (31.7%) were the most effective drugs for routine use among the P. aeruginosa strains studied (34). Similarly, Hosu et al. reported 16.7 and 22.2% resitance for amikacin and ciprofloxacin respectively (13). In accordance with our findings, Mobaraki in Iran reported increased resistance for both amikacin and ciprofloxacin from the year 2007 to 2014 (35). Furthermore, Poonsuk et al. (36) showed high-level resistance of P. aeruginosa isolates to amikacin (92.1%), and ciprofloxacin (95%) (37). Resistance to such antibiotics increased in comparison to previous studies, which could be attributed to variations in antibiotic usage (3).
5.1. Conclusions
This discovery revealed differences in the virulence profiles and antibiotic resistance of P. aeruginosa strains isolated from animal and human samples, demonstrating the adaptive ability of this bacterial species. According to the current study, wound patients are more likely to get infections than other patients. In conclusion, isolated P. aeruginosa isolates exhibit several number of exoA and lasB virulence genes which may play an important role in infections caused by P. aeruginosa. It was observed that high percentages of resistance to amikacin and low percentages of resistance to ciprofloxacin among all the examined antibiotics which is cause for concern. As a result, antibiotic consumption and management are critical for reducing resistance and effectively treating infections caused by the P. aeruginosa bacterium.
References
-
1.
El Shamy AA, Zakaria Z, Tolba MM, Salah Eldin N, Rabea AT, Tawfick MM, et al. AmpC beta-Lactamase Variable Expression in Common Multidrug-Resistant Nosocomial Bacterial Pathogens from a Tertiary Hospital in Cairo, Egypt. Int J Microbiol. 2021;2021:6633888. [PubMed ID: 33854549]. [PubMed Central ID: PMC8021464]. https://doi.org/10.1155/2021/6633888.
-
2.
Benie CK, Dadie A, Guessennd N, N'Gbesso-Kouadio NA, Kouame ND, N'Golo D C, et al. Characterization of Virulence Potential of Pseudomonas Aeruginosa Isolated from Bovine Meat, Fresh Fish, and Smoked Fish. Eur J Microbiol Immunol (Bp). 2017;7(1):55-64. [PubMed ID: 28386471]. [PubMed Central ID: PMC5372481]. https://doi.org/10.1556/1886.2016.00039.
-
3.
Hu H, Manos J. Pulsed-field gel electrophoresis of Pseudomonas aeruginosa. Methods Mol Biol. 2015;1301:157-70. [PubMed ID: 25862056]. https://doi.org/10.1007/978-1-4939-2599-5_14.
-
4.
Rocha AJ, Barsottini MRDO, Rocha RR, Laurindo MV, Moraes FLLD, Rocha SLD. Pseudomonas Aeruginosa: Virulence Factors and Antibiotic Resistance Genes. Braz Arch Biol Technol. 2019;62. https://doi.org/10.1590/1678-4324-2019180503.
-
5.
Chand Y, Khanal S, Panta OP, Shrestha D, Khadka DK, Poudel P. Prevalence of some virulence genes and antibiotic susceptibility pattern of Pseudomonas aeruginosa isolated from different clinical specimens. ResearchSquare. 2020;Preprint. https://doi.org/10.21203/rs.3.rs-24044/v1.
-
6.
Saadoon ZS, Zghair ZR. Molecular Detection of Pseudomonas aeruginosa by Using Algd, Plch and Lasb Genes and Pathological Study of the Virulent Isolate from Human Blood. Plant Archives. 2019;19(2):1633-9.
-
7.
Pillar CM, Hobden JA. Pseudomonas aeruginosa exotoxin A and keratitis in mice. Invest Ophthalmol Vis Sci. 2002;43(5):1437-44. [PubMed ID: 11980858].
-
8.
Yousefi-Avarvand A, Khashei R, Sedigh Ebrahim-Saraie H, Emami A, Zomorodian K, Motamedifar M. The Frequency of Exotoxin A and Exoenzymes S and U Genes Among Clinical Isolates of Pseudomonas aeruginosa in Shiraz, Iran. Int J Mol Cell Med. 2015;4(3):167-73. [PubMed ID: 26629485]. [PubMed Central ID: PMC4644528].
-
9.
Khattab MA, Nour MS, ElSheshtawy NM. Genetic Identification of Pseudomonas aeruginosa Virulence Genes among Different Isolates. J Microb Biochem Technol. 2015;7(5):274-7. https://doi.org/10.4172/1948-5948.1000224.
-
10.
Zarei O, Shokoohizadeh L, Hossainpour H, Alikhani MY. Molecular analysis of Pseudomonas aeruginosa isolated from clinical, environmental and cockroach sources by ERIC-PCR. BMC Res Notes. 2018;11(1):668. [PubMed ID: 30219108]. [PubMed Central ID: PMC6139122]. https://doi.org/10.1186/s13104-018-3765-z.
-
11.
Mohanty S, Baliyarsingh B, Kumar Nayak S. Antimicrobial Resistance in Pseudomonas aeruginosa: A Concise Review. In: Mares M, Erin Lim SH, Lai K, Cristina R, editors. Antimicrobial Resistance. London, UK: IntechOpen; 2021. https://doi.org/10.5772/intechopen.88706.
-
12.
Bajpai V, Govindaswamy A, Khurana S, Batra P, Aravinda A, Katoch O, et al. Phenotypic & genotypic profile of antimicrobial resistance in Pseudomonas species in hospitalized patients. Indian J Med Res. 2019;149(2):216-21. [PubMed ID: 31219086]. [PubMed Central ID: PMC6563739]. https://doi.org/10.4103/ijmr.IJMR_1_18.
-
13.
Hosu MC, Vasaikar S, Okuthe GE, Apalata T. Molecular Detection of Antibiotic-Resistant Genes in Pseudomonas aeruginosa from Nonclinical Environment: Public Health Implications in Mthatha, Eastern Cape Province, South Africa. Int J Microbiol. 2021;2021:8861074. [PubMed ID: 33519937]. [PubMed Central ID: PMC7817236]. https://doi.org/10.1155/2021/8861074.
-
14.
Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177-92. [PubMed ID: 30500353]. https://doi.org/10.1016/j.biotechadv.2018.11.013.
-
15.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne, USA: Clinical and Laboratory Standards Institute; 2011.
-
16.
J. Al-zubaidy B, R. Abdulrahman T, A. Ahmed W. Detection of Exo A Gene in Pseudomonas aeruginosa from Human and Dogs Using Polymerase Chain Reaction. IOSR J Pharm Biol Sci. 2014;9(5):38-41. https://doi.org/10.9790/3008-09563841.
-
17.
Mokhtari K, Amini K. Identification of virulence genes in Pseudomonas aeruginosa isolated from human and animal samples by multiplex-PCR and their antibiotic resistance pattern. Iran J Med Mic. 2019;13(4):294-301. https://doi.org/10.30699/ijmm.13.4.294.
-
18.
Golle A, Janezic S, Rupnik M. Low overlap between carbapenem resistant Pseudomonas aeruginosa genotypes isolated from hospitalized patients and wastewater treatment plants. PLoS One. 2017;12(10). e0186736. [PubMed ID: 29049368]. [PubMed Central ID: PMC5648238]. https://doi.org/10.1371/journal.pone.0186736.
-
19.
Heinzelmann M, Scott M, Lam T. Factors predisposing to bacterial invasion and infection. Am J Surg. 2002;183(2):179-90. https://doi.org/10.1016/s0002-9610(01)00866-2.
-
20.
Dursun A, Özsoylu S, Kılıç H, Ulu Kılıç A, Akyıldız BN. Antibiotic Susceptibilities of Pseudomonas aeruginosa, Klebsiella pneumoniae and Acinetobacter baumannii Strains Isolated from Patients in the Pediatric Intensive Care Unit. Turk J Intens Care. 2018;16(3):109-14. https://doi.org/10.4274/tybd.63825.
-
21.
Ranjan KP, Ranjan N, Bansal SK, Arora DR. Prevalence of Pseudomonas aeruginosa in post-operative wound infection in a referral hospital in Haryana, India. J Lab Physicians. 2010;2(2):74-7. [PubMed ID: 21346900]. [PubMed Central ID: PMC3040092]. https://doi.org/10.4103/0974-2727.72153.
-
22.
Faraji F, Mahzounieh M, Ebrahimi A, Fallah F, Teymournejad O, Lajevardi B. Molecular detection of virulence genes in Pseudomonas aeruginosa isolated from children with Cystic Fibrosis and burn wounds in Iran. Microb Pathog. 2016;99:1-4. [PubMed ID: 27457974]. https://doi.org/10.1016/j.micpath.2016.07.013.
-
23.
O. M. Al-Dahmoshi H, S. Al-Khafaji N, Abdulzahra Jeyad A, Khaleel Shareef H, F. Al-Jebori R. Molecular Detection of Some Virulence Traits among Pseudomonas aeruginosa Isolates, Hilla-Iraq. Biomed Pharmacol J. 2018;11(2):835-42. https://doi.org/10.13005/bpj/1439.
-
24.
Aljebory IS. Genetic Detection and Identification of Some Virulence Factors Genes Among Pseudomonas aeruginosa Samples in Kirkuk province-Iraq. Int J Pharm Qual Assur. 2019;10(3). https://doi.org/10.25258/ijpqa.10.3.8.
-
25.
Amirmozafari N, Mehrabadi JF, Habibi A. Association of the exotoxin A and exoenzyme S with antimicrobial resistance in Pseudomonas aeruginosa strains. Arch Iran Med. 2016;19(5):353-8.
-
26.
Neamah AA. Molecular Detection of virulence factor genes in Pseudomonas aeruginosa isolated from human and animals in Diwaniya province. Kufa J Vet Med Sic. 2017;8:218-30.
-
27.
Dekhil KR. Molecular characterization and phylogenetic analysis of Pseudomonas aeruginosa isolated from active chronic otitis media. Int J Res Pharm Sci. 2017;8(3):389-96.
-
28.
Sabharwal N, Dhall S, Chhibber S, Harjai K. Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. Int J Mol Epidemiol Genet. 2014;5(3):125. [PubMed ID: 25379131]. [PubMed Central ID: PMC4214259].
-
29.
Freschi L, Vincent AT, Jeukens J, Emond-Rheault JG, Kukavica-Ibrulj I, Dupont MJ, et al. The Pseudomonas aeruginosa Pan-Genome Provides New Insights on Its Population Structure, Horizontal Gene Transfer, and Pathogenicity. Genome Biol Evol. 2019;11(1):109-20. [PubMed ID: 30496396]. [PubMed Central ID: PMC6328365]. https://doi.org/10.1093/gbe/evy259.
-
30.
Nikbin VS, Aslani MM, Sharafi Z, Hashemipour M, Shahcheraghi F, Ebrahimipour GH. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran J Microbiol. 2012;4(3):118-23. [PubMed ID: 23066485]. [PubMed Central ID: PMC3465536].
-
31.
Javiya VA, Ghatak SB, Patel KR, Patel JA. Antibiotic susceptibility patterns of Pseudomonas aeruginosa at a tertiary care hospital in Gujarat, India. Indian J Pharmacol. 2008;40(5):230-4. [PubMed ID: 20040963]. [PubMed Central ID: PMC2792624]. https://doi.org/10.4103/0253-7613.44156.
-
32.
Gales AC, Jones RN, Turnidge J, Rennie R, Ramphal R. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32 Suppl 2:S146-55. [PubMed ID: 11320454]. https://doi.org/10.1086/320186.
-
33.
Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R. Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals. 2014;42(1):1-7. [PubMed ID: 24309094]. https://doi.org/10.1016/j.biologicals.2013.11.001.
-
34.
Drenkard E. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microb Infec. 2003;5(13):1213-9. https://doi.org/10.1016/j.micinf.2003.08.009.
-
35.
Dou Y, Huan J, Guo F, Zhou Z, Shi Y. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J Int Med Res. 2017;45(3):1124-37. [PubMed ID: 28443385]. [PubMed Central ID: PMC5536433]. https://doi.org/10.1177/0300060517703573.
-
36.
Poonsuk K, Tribuddharat C, Chuanchuen R. Class 1 integrons in Pseudomonas aeruginosa and Acinetobacter baumannii isolated from clinical isolates. Southeast Asian J Trop Med Public Health. 2012;43(2):376.
-
37.
Bellés Bellés A. Class 1 integrons from Pseudomonas aeruginosa isolates from lower tract respiratory samples. Berlin, Germany: Morressier; 2016. https://doi.org/10.26226/morressier.56d6be75d462b80296c976e8.