Abstract
Background:
Gliosarcoma (GS) is a rare primary neoplasm of the central nervous system. It is a subtype of glioblastoma and has a biphasic pattern consisting of glial and malignant mesenchymal elements. Its onset is between the fourth and sixth decade of life.Objectives:
Since protein - protein interaction (PPI) network analysis can provide useful information about molecular aspects of diseases, the aim of this study is GS protein analysis via PPI network and gene ontology assessment.Methods:
The related genes to GS were gathered from STRING DB and organized in the interacted network by Cytoscape software version 3.6.0. The network was analyzed based on topological parameters and the central nodes were introduced. The significant clusters were identified by ClusterONE and the cluster included more key genes enriched via gene ontology by ClueGO.Results:
Nine crucial genes including TP53, EGFR, PTEN, EGR1, VEGFA, HSP90AA1, IL2, KNG1, and HSP90AB1 were introduced as related key genes to GS. Two significant clusters contain most of central genes. Twenty - one elements of cluster - 1, which included 7 key genes, were enriched via gen ontology and 115 related terms were determined and discussed.Conclusions:
The nine introduced central genes may play main roles in pathology of GS. However, experimental investigation is proposed to validate the findings.Keywords
1. Background
One of the rare primary neoplasms of the central nervous system is gliosarcoma (GS), a subtype of glioblastoma (GBM) (1). GS was first reported by Strobe in 1895. It has a biphasic pattern consisting of glial (anaplastic astrocytes) and malignant mesenchymal elements. However, the monoclonal or biclonal origin of GS biphasic nature is still subject to debate (2). The onset of GS, as a rare neoplasm, is between the fourth and sixth decade of life (3), and the male/female ratio of GS is 1.8/1 (4). Treatment consists of surgical resection of the tumor followed by external radiotherapy or chemotherapy in some cases (4, 5). Authors represented different genes involved in the disease as p53 mutant expression (6, 7). Glial fibrillary acidic protein also reported as an diagnostic protein embedded in glioblastoma cells (8, 9). Scientists assessed the genetic profile of GS with mutation in P53, PTEN, and deletion of P16 with CDK4 amplification (10). Douglas et al. from Stanford compared systematic genes variations in 60 different cancer cell lines. Their article published in Genetics Nature represented common genes involved in gliosarcoma and other cancer cells (11). On the other hand, the mesenchymal component of GSs can present differentiation along several lineages as fibroblasts and chondroblasts, etc. (12). Investigation indicates that EGFR amplification is much lower in GS than GMB (13). Identifying the genes and proteins involved in the development of GS or the other types of cancer can effectively determine their treatment (14). From the perspective of systems biology, the connection between proteins involved in the disease is important (15). PPI network analysis of diseases has attracted attention of medical and biological scientist. In this approach, examination of the interaction between genes involved in the disease could lead us to improve the diagnosis and treatment of patients (16-20). In PPI network analysis, the related genes to the disease are gathered and organized in an integrative structure as a interactome (16, 21). The assessment of topological properties of the network, including central parameters such as degree and betweenness centrality provides useful information about molecular mechanism of disease onset and pathology (22-24). Introducing selected genes among large number of query genes can lead to specific biomarker panel related to the disease (25). In this study, we aim at identifying and analyzing genes interaction involved in GS disease. It may be conducted to introduce a biomarker panel related to GS.
2. Methods
A number of 200 genes related to gliosurcoma were searched from diseas query of String data bank (String is one of Cytoscape applications), but only 106 genes were introduced. The genes were analyzed for network construction by Cytoscape software version 3.6.0 (26). The main connected component of PPI network includes 78 nodes and 269 undircted edges were layout by degree values. The top 20% of the nodes based on degree values were selected as hub genes. The bottleneck nodes were identified considering top 20% of the nodes based on betweenness centerality. Similarly, top 20% of the nodes based on closeness centerality and stress values were selected as centeral genes. The hub nodes, which are bottleneck node, are introuced as hub - bottlenck genes. The nodes that were hub - bottleneck genes and their closeness and stress ranks were less than 10 and selected as crucial nodes. The mian connected component (the constructed network) was analyzed by Cluster ONE plugin of Cytoscape software (27). The significant clusters were introduced and discussed. Finaly, the main identified clusters were analyzed by ClueGO (28). The gene onthology finding were assested and categorized in the related groups.
3. Results
The 106 related genes of gliosarcoma for human species were retrieved from disease query of String database (confides 0.40 score). The genes were organized in a PPI network, including a main connected component (a network constructed with 78 nodes and 296 undirected edges) and 28 isolated nodes. The network is presented in Figure 1. The network was analyzed and for more resolution, the 20% of top nodes (equal to 16 nodes) based on degree values were selected as hub - genes. Similarly, 16 top nodes based on betweenness values were identified as bottleneck - genes. Two separated groups (each of them includes 16 top genes, corresponding to the values of closeness centrality and stress) were determined as central nodes. Therefore, 64 nodes were organized in 4 groups. If an individual node was not hub or bottleneck genes and was not found in the one of the other two groups, it would be omitted from study. Finally, 9 crucial genes of the analyzed network were identified and presented in Table 1. Connections between the crucial genes were illustrated in Figure 2 as a sub network. This sub network was resulted via the deletion of the other elements of gliosarcoma network. Since members of a cluster (cluster plays a role as an integrative part of a network) are involved in closed functional terms, cluster analysis of the network limited to the presence of at least 10 nodes in a cluster was performed. The cluster analysis revealed that there are 2 clusters (cluster 1 and 2) characterized by different quality and P value related to the constructed network. These clusters in interaction with the other parts of the network are presented in the Figures 3 and 4. Since cluster - 1 contains more crucial genes, this cluster was analyzed via gene ontology method. Gene ontology for the nodes of cluster - 1 indicates that 115 biological terms including biological processes, cellular components, molecular function, and biochemical pathways (from KEGG data base) are related to this cluster (Figure 5). The term was included if at least 2 genes and 5% gene/term contribution were considered. The analysis of a large number of the terms (115 terms) is a difficult process; therefore, as it is shown in Figure 6, the terms are organized in 5 groups.
PPI network of gliosarcoma is constructed with 78 nodes and 296 undirected edges. The network is layout by degree value. The bigger size of a node corresponds to bigger value of degree. Color from gray to blue refers to increment of degree value.
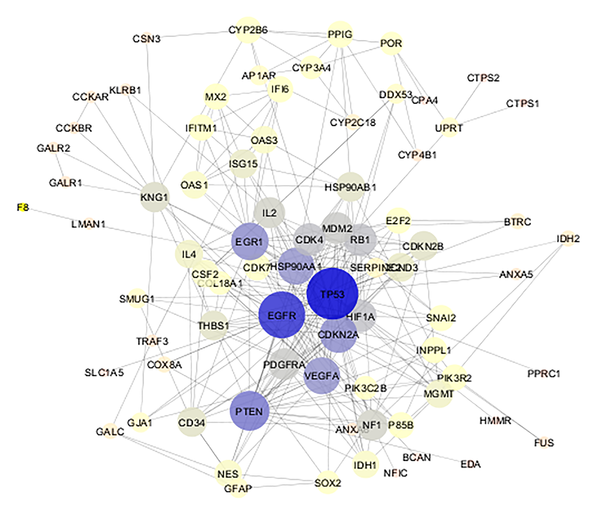
The 9 crucial nodes related to gliosarcoma are presented. All nodes (except HSP90AB1) are hub - bottleneck genes. HSP90AB1 was not included in hub - genes, but is identified as a bottleneck node. D, BC, CC, S, and DS are degree, betweenness centrality, closeness centrality, stress, and disease sore, respectively.
| R | Name | Description | D | BC | CC | S | DS |
|---|---|---|---|---|---|---|---|
| 1 | TP53 | Tumor protein p53 | 42 | 0.31 | 0.63 | 4896 | 2.58 |
| 2 | EGFR | Epidermal growth factor receptor | 35 | 0.26 | 0.61 | 4104 | 1.51 |
| 3 | PTEN | Phosphatase and tensin homolog | 25 | 0.07 | 0.53 | 1448 | 1.12 |
| 4 | EGR1 | Early growth response 1 | 22 | 0.10 | 0.54 | 2044 | 0.56 |
| 5 | VEGFA | Vascular endothelial growth factor A | 21 | 0.03 | 0.53 | 884 | 0.61 |
| 6 | HSP90AA1 | Heat shock protein 90kDa alpha (cytosolic), class A member 1 | 21 | 0.13 | 0.55 | 3054 | 0.63 |
| 7 | IL2 | Interleukin 2 | 13 | 0.02 | 0.48 | 566 | 1.33 |
| 8 | KNG1 | Kininogen 1 | 12 | 0.12 | 0.48 | 2036 | 0.55 |
| 9 | HSP90AB1 | Heat shock protein 90kDa alpha (cytosolic), class B member 1 | 11 | 0.3 | 0.49 | 566 | 1.46 |
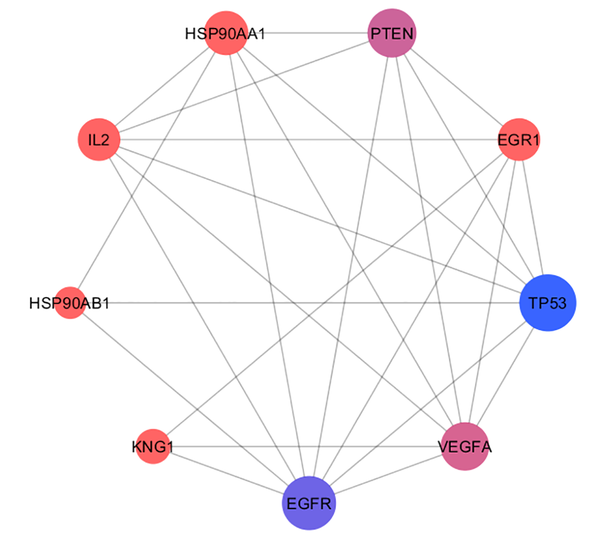
Connections between the crucial genes of gliosarcoma PPI network. The nodes are layout based on degree value. The bigger size refers to higher degree value. Color from orange to blue is corresponding to degree value increment.
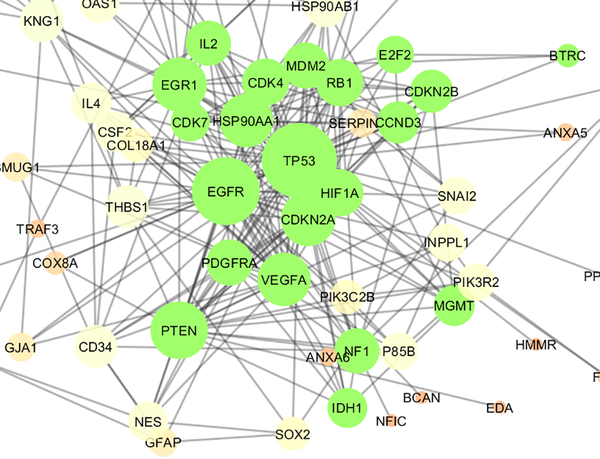
Cluster - 1 is constructed of 21 nodes (shown in green color) including 7 top genes of Table 1. The nodes are layout by degree value. P value, Quality, and Density were equal to 10- 4, 0.571, and 0.543 respectively.
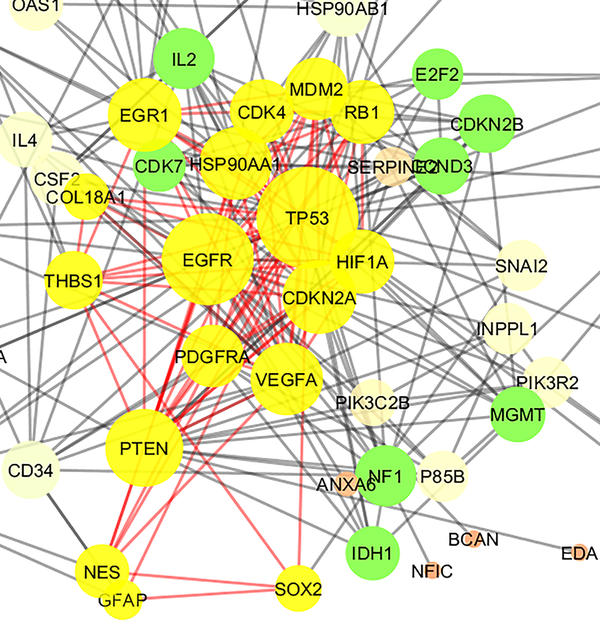
Cluster - 2 is constructed of 17 nodes (shown in yellow color) including 6 top genes of Table 1. The nodes are layout by degree value. P value, Quality, and Density were equal to 0.034, 0.387, and 0.603, respectively.
Gene ontology result for the nodes of cluster - 1, including biological processes, cellular components, molecular function, and biochemical pathways (from KEGG database). The term was included if at least 2 genes and 5% gene/term contribution were considered.
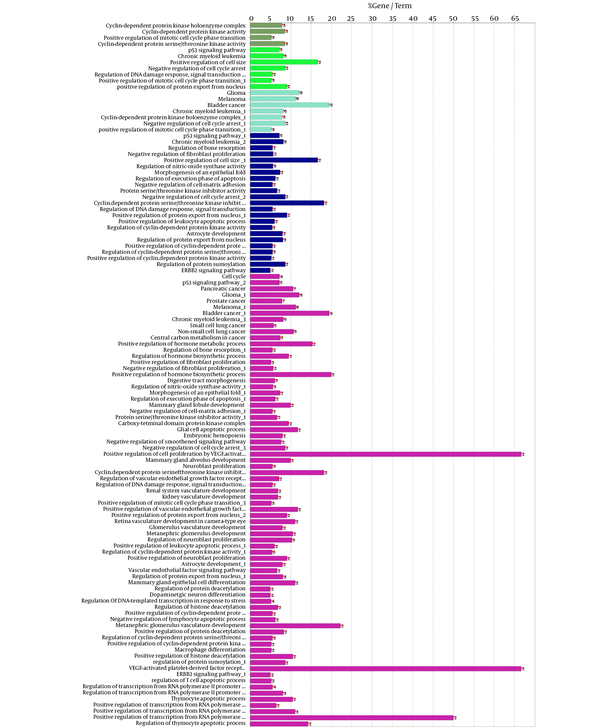
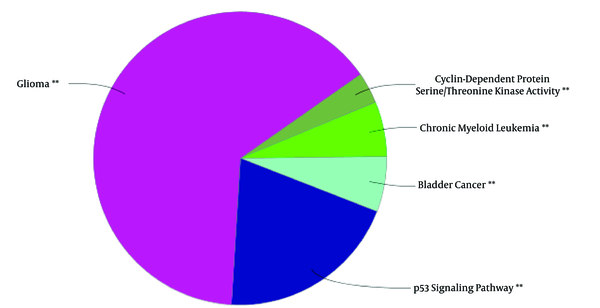
The represented 115 terms in figure 4 are grouped. Term P values and group P values are less than 10- 3.
4. Discussion
Gliosarcoma is a disease, which is studied by several molecular approaches such as genetics molecular biology and metabolite analysis (10, 29, 30). Therefore, there are a number of genes that are related to this disease. It is an essential need to rank and screen the genes for introducing the important elements among them. PPI network analysis is the right approach in this regard. The constructed network in this study contains 78 nodes and 296 edges. It seems that compared to other diseases PPI network, this network is a small network. Topology analysis of the network based on central parameters revealed that there are 9 crucial genes related to GS. Here, we tried to represent an overview of the significant roles of identified crucial genes of network in GC pathogenesis and development.
As it is known, TP53 is the most famous gene related to cancers. Rarely, it is discussed about the cancer without TP53 participation. TP53 mutations accompanied with the other genes such as CDKN2A is reported in GS and it is discussed in details (31). In the another report, the role of TP53 mutation beside CD34 in the GS patients was studied and its significant role in the disease was emphasized (32). In addition, the amplification of EGFR in correlation with PTEN in the glioblastoma and GS patients were studied and confirmed (33). Phosphatidyl - inositol - 3’ - kinase (Pi3k) - dependent activation of protein kinase B (Akt) signaling is the inhibitory role of PTEN as a tumor suppressor gene (34). Investigation indicates that EGFR over expression, occurrence mutations in PTEN, and P16 deletions accompanied with MDM2 amplification are the typical events in this disease (35). EGR1 is responsible for synaptic plasticity and neuronal activity in both physiological and pathological conditions (36). A study indicates that increment of VEGF - A and TGFβ2 signaling invokes gene expression changes related to glioblastoma vessels (37).
The significant role of HSP90AA1 and HSP90AB1 in several cancer types such as lung cancer, ovarian cancer, and GS is detected and discussed (38-40). The attenuation role of IL2 in growth of glioma and brain edema in rats is assessed and discussed in details (41). Down regulation of KNG1 in bladder and gastric cancers is studied and confirmed (42, 43). As it was shown, there is an exact relationship between expression changes of the nine introduced key genes and cancer diseases especially GS.
Consequently, the represented crucial genes probably are right related genes to GS.
A cluster is a combination of elements with tight relationships. In this study, a cluster was introduced containing the most number of the important genes, which is a novel finding. The cluster analysis identified 2 significant clusters. Cluster - 1 includes 21 nodes, in which 7 were common with the key genes. Similarly, cluster - 2 includes 17 nodes and 6 common key genes. The finding indicates that cluster-1 and cluster - 2 are 78% and 67% common considering key genes, respectively. Based on the findings, it can be concluded that cluster - 1 is a main structural component of the network. Thus, gene ontology enrichment of elements of cluster - 1 can lead to introduce the related terms of GS. A number of 115 terms were recognized, in which several terms are characterized by high percentage of attribution such as the VEGF involved terms. On the other hand, there are several terms, which are specified by the presence of more genes in the terms such as glioma, melanoma, cell cycle, and bladder cancers. Since the interpretation of a large number of the terms is difficult, the terms were categorized in 5 groups. The main group is determined as glioma, which indicates that our analysis was conducted in an appropriate way. The second important group is p53 signaling pathway. There is evidence about correlation between p53 pathway and glioma (44). The other groups were involved in different malignancies.
4.1. Conclusion
The 9 determined key genes may be the central core of GS and play major roles in pathology of disease. The validation of findings via experimental investigation is useful for the approval, tool.
Acknowledgements
References
-
1.
Singh G, Das KK, Sharma P, Guruprasad B, Jaiswal S, Mehrotra A, et al. Cerebral gliosarcoma: Analysis of 16 patients and review of literature. Asian J Neurosurg. 2015;10(3):195-202. [PubMed ID: 26396606]. [PubMed Central ID: PMC4553731]. https://doi.org/10.4103/1793-5482.161173.
-
2.
Pardo J, Murcia M, Garcia F, Alvarado A. Gliosarcoma: A rare primary CNS tumor. Presentation of two cases. Rep Pract Oncol Radiother. 2010;15(4):98-102. [PubMed ID: 24376932]. [PubMed Central ID: PMC3863303]. https://doi.org/10.1016/j.rpor.2010.05.003.
-
3.
Little JA, Shanoff HM, Roe RD, Csima A, Yano R. Studies of Male Survivors of Myocardial Infarction. Iv. Serum Lipids and Five-Year Survival. Circulation. 1965;31:854-62. [PubMed ID: 14297519].
-
4.
Hattab EM, Harris BT. Molecular Pathology of the Central Nervous System. Molecular Genetic Pathology. Springer; 2008. p. 497-532.
-
5.
Newton HB, Otero JJ. Overview of epidemiology, pathology, and treatment of primary brain tumors. Epilepsy and Brain Tumors. Elsevier; 2015. p. 11-28.
-
6.
Cho SY, Park C, Na D, Han JY, Lee J, Park OK, et al. High prevalence of TP53 mutations is associated with poor survival and an EMT signature in gliosarcoma patients. Exp Mol Med. 2017;49(4). e317. [PubMed ID: 28408749]. [PubMed Central ID: PMC5420801]. https://doi.org/10.1038/emm.2017.9.
-
7.
Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829-48. [PubMed ID: 25943888]. https://doi.org/10.1007/s00401-015-1432-1.
-
8.
Behling F, Barrantes-Freer A, Skardelly M, Nieser M, Christians A, Stockhammer F, et al. Frequency of BRAF V600E mutations in 969 central nervous system neoplasms. Diagn Pathol. 2016;11(1):55. [PubMed ID: 27350555]. [PubMed Central ID: PMC4924254]. https://doi.org/10.1186/s13000-016-0506-2.
-
9.
Nsir AB, Thai Q-A, Kassar AZ, Said IB, Jemel H. Primary cerebellar gliosarcoma with extracranial metastases: an orphan differential diagnosis. World Neurosurg. 2015;84(6):2076. e13-7.
-
10.
Reis RM, Konu-Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H. Genetic profile of gliosarcomas. Am J Pathol. 2000;156(2):425-32. [PubMed ID: 10666371]. [PubMed Central ID: PMC1850048]. https://doi.org/10.1016/S0002-9440(10)64746-3.
-
11.
Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227-35. [PubMed ID: 10700174]. https://doi.org/10.1038/73432.
-
12.
deCarvalho AC, Nelson K, Lemke N, Lehman NL, Arbab AS, Kalkanis S, et al. Gliosarcoma stem cells undergo glial and mesenchymal differentiation in vivo. Stem Cells. 2010;28(2):181-90. [PubMed ID: 19937755]. [PubMed Central ID: PMC2873790]. https://doi.org/10.1002/stem.264.
-
13.
Miller CR, Perry A. Glioblastoma. Arch Pathol Lab Med. 2007;131(3):397-406. [PubMed ID: 17516742]. https://doi.org/10.1043/1543-2165(2007)131[397:G]2.0.CO;2.
-
14.
Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, Goliaei B, Peyvandi AA. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014;7(1):17-31. [PubMed ID: 25436094]. [PubMed Central ID: PMC4017556].
-
15.
Karbalaei R, Piran M, Rezaei-Tavirani M, Asadzadeh-Aghdaei H, Heidari MH. A systems biology analysis protein-protein interaction of NASH and IBD based on comprehensive gene information. Gastroenterol Hepatol Bed Bench. 2017;10(3):194-201. [PubMed ID: 29118935]. [PubMed Central ID: PMC5660269].
-
16.
Zali H, Rezaei Tavirani M. Meningioma protein-protein interaction network. Arch Iran Med. 2014;17(4):262-72. [PubMed ID: 24724603].
-
17.
Safari-Alighiarloo N, Rezaei-Tavirani M, Taghizadeh M, Tabatabaei SM, Namaki S. Network-based analysis of differentially expressed genes in cerebrospinal fluid (CSF) and blood reveals new candidate genes for multiple sclerosis. PeerJ. 2016;4. e2775. [PubMed ID: 28028462]. [PubMed Central ID: PMC5183126]. https://doi.org/10.7717/peerj.2775.
-
18.
Peyvandi AA, Khoshsirat S, Safaei A, Rezaei-Tavirani M, Azodi-Zamanian M. Interactome Analysis of 11-Dehydrosinulariolide-Treated Oral Carcinoma Cell Lines Such as Ca9-22 and CAL-27 and Melanoma Cell Line. Int J Canc Manag. 2017;10(7). https://doi.org/10.5812/ijcm.10096.
-
19.
Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25(10):1119-26. [PubMed ID: 17921997]. https://doi.org/10.1038/nbt1338.
-
20.
Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput Biol. 2010;6(1). e1000641. [PubMed ID: 20090828]. [PubMed Central ID: PMC2797085]. https://doi.org/10.1371/journal.pcbi.1000641.
-
21.
Abbaszadeh HA, Peyvandi AA, Sadeghi Y, Safaei A, Zamanian-Azodi M, Khoramgah MS, et al. Er:YAG Laser and Cyclosporin A Effect on Cell Cycle Regulation of Human Gingival Fibroblast Cells. J Lasers Med Sci. 2017;8(3):143-9. [PubMed ID: 29123635]. [PubMed Central ID: PMC5662504]. https://doi.org/10.15171/jlms.2017.26.
-
22.
Rezaei-Tavirani M, Rezaei-Tavirani M, Mansouri V, Mahdavi SM, Valizadeh R, Rostami-Nejad M, et al. Introducing crucial protein panel of gastric adenocarcinoma disease. Gastroenterol Hepatol Bed Bench. 2017;10(1):21-8. [PubMed ID: 28331560]. [PubMed Central ID: PMC5346820].
-
23.
Safaei A, Rezaei Tavirani M, Zamanian Azodi M, Lashay A, Mohammadi SF, Ghasemi Broumand M, et al. Diabetic Retinopathy and Laser Therapy in Rats: A Protein-Protein Interaction Network Analysis. J Lasers Med Sci. 2017;8(Suppl 1):S20-1. [PubMed ID: 29071030]. [PubMed Central ID: PMC5642173]. https://doi.org/10.15171/jlms.2017.s4.
-
24.
Manzoni C, Ferrari R, Kia D, Lovering RC, Hardy J, Lewis PA. Protein Network Analysis to Prioritize Candidate Genes and Pathways for Sporadic Disease: A Comparison between Frontotemporal Dementia and Parkinson’s Disease. Alzheimer's & Dementia. 2017;13(7):P555. https://doi.org/10.1016/j.jalz.2017.07.156.
-
25.
Rezaei Tavirani M, Ok HF, Zamanian Azodi M, Rezaei Tavirani M. Duchenne Muscular Dystrophy (DMD) Protein-Protein Interaction Mapping. Iran J Child Neurol. 2017;11(4):7-14. [PubMed ID: 29201118]. [PubMed Central ID: PMC5703623].
-
26.
Khayer N, Zamanian-Azodi M, Mansouri V, Ghassemi-Broumand M, Rezaei-Tavirani M, Heidari MH, et al. Oral squamous cell cancer protein-protein interaction network interpretation in comparison to esophageal adenocarcinoma. Gastroenterol Hepatol Bed Bench. 2017;10(2):118-24. [PubMed ID: 28744358]. [PubMed Central ID: PMC5495899].
-
27.
Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi-Nejad A, Rostami-Nejad M, Rahmatirad S. Protein Clustering and Interactome Analysis in Parkinson and Alzheimer's Diseases. Arch Iran Med. 2016;19(2):101-9. [PubMed ID: 26838080].
-
28.
Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Rezaei Tavirani M, Seyyedi SS. Protein-Protein Interaction Network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed Bench. 2015;8(3):215-24. [PubMed ID: 26328044]. [PubMed Central ID: PMC4553162].
-
29.
Huttner A. Overview of primary brain tumors: pathologic classification, epidemiology, molecular biology, and prognostic markers. Hematol Oncol Clin North Am. 2012;26(4):715-32. [PubMed ID: 22794280]. https://doi.org/10.1016/j.hoc.2012.05.004.
-
30.
Warnick RE, Pietronigro DD, McBride DQ, Flamm ES. In vivo metabolism of radiolabeled putrescine in gliomas: implications for positron emission tomography of brain tumors. Neurosurgery. 1988;23(4):464-9. [PubMed ID: 3264387].
-
31.
Deb P, Sharma MC, Chander B, Mahapatra AK, Sarkar C. Giant cell glioblastoma multiforme: report of a case with prolonged survival and transformation to gliosarcoma. Childs Nerv Syst. 2006;22(3):314-9. [PubMed ID: 16133270]. https://doi.org/10.1007/s00381-005-1239-9.
-
32.
Machuca TN, Prevedello DM, Pope LZ, Haratz SS, Araujo JC, Torres LF. Gliosarcoma: report of four cases with immunohistochemical findings. Arq Neuropsiquiatr. 2004;62(3A):608-12. [PubMed ID: 15334217].
-
33.
Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27(4):579-84. [PubMed ID: 19075262]. [PubMed Central ID: PMC2645859]. https://doi.org/10.1200/JCO.2008.18.9639.
-
34.
Knobbe CB, Reifenberger G. Genetic alterations and aberrant expression of genes related to the phosphatidyl-inositol-3'-kinase/protein kinase B (Akt) signal transduction pathway in glioblastomas. Brain Pathol. 2003;13(4):507-18. [PubMed ID: 14655756].
-
35.
Kleihues P, Ohgaki H. Phenotype vs genotype in the evolution of astrocytic brain tumors. Toxicol Pathol. 2000;28(1):164-70. [PubMed ID: 10669004]. https://doi.org/10.1177/019262330002800121.
-
36.
Duclot F, Kabbaj M. The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Front Behav Neurosci. 2017;11:35. [PubMed ID: 28321184]. [PubMed Central ID: PMC5337695]. https://doi.org/10.3389/fnbeh.2017.00035.
-
37.
Dieterich LC, Mellberg S, Langenkamp E, Zhang L, Zieba A, Salomaki H, et al. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFbeta2 in vascular abnormalization. J Pathol. 2012;228(3):378-90. [PubMed ID: 22786655]. https://doi.org/10.1002/path.4072.
-
38.
Coskunpinar E, Akkaya N, Yildiz P, Oltulu YM, Aynaci E, Isbir T, et al. The significance of HSP90AA1, HSP90AB1 and HSP90B1 gene polymorphisms in a Turkish population with non-small cell lung cancer. Anticancer Res. 2014;34(2):753-7. [PubMed ID: 24511009].
-
39.
Chu SH, Liu YW, Zhang L, Liu B, Li L, Shi JZ, et al. Regulation of survival and chemoresistance by HSP90AA1 in ovarian cancer SKOV3 cells. Mol Biol Rep. 2013;40(1):1-6. [PubMed ID: 23135731]. https://doi.org/10.1007/s11033-012-1930-3.
-
40.
Gorska M, Marino Gammazza A, Zmijewski MA, Campanella C, Cappello F, Wasiewicz T, et al. Geldanamycin-induced osteosarcoma cell death is associated with hyperacetylation and loss of mitochondrial pool of heat shock protein 60 (hsp60). PLoS One. 2013;8(8). e71135. [PubMed ID: 24015183]. [PubMed Central ID: PMC3756027]. https://doi.org/10.1371/journal.pone.0071135.
-
41.
Tjuvajev J, Gansbacher B, Desai R, Beattie B, Kaplitt M, Matei C, et al. RG-2 glioma growth attenuation and severe brain edema caused by local production of interleukin-2 and interferon-gamma. Cancer Res. 1995;55(9):1902-10. [PubMed ID: 7728757].
-
42.
Li F, Chen DN, He CW, Zhou Y, Olkkonen VM, He N, et al. Identification of urinary Gc-globulin as a novel biomarker for bladder cancer by two-dimensional fluorescent differential gel electrophoresis (2D-DIGE). J Proteomics. 2012;77:225-36. [PubMed ID: 22986152]. https://doi.org/10.1016/j.jprot.2012.09.002.
-
43.
Fan L-Q, Li Y, Chen M-L, Zhao Q, Tan B-B, Wang D, et al. AB27. Proteomics research about effect of ZNF139 knockdown with RNA interference on serum of orthotopic transplantation nude mouse with gastric cancer. Translational Gastrointestinal Cancer. 2013.
-
44.
Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60(2):417-24. [PubMed ID: 10667596].
