Abstract
Background:
Worldwide, colorectal cancer is the fourth most common cause of cancer-related deaths. Infectious agents have long been associated with development of gastrointestinal malignancies including colorectal cancer. Therefore, the aim of this study was to detect Streptococcus gallolyticus subsp. gallolyticus and Helicobacter pylori in colorectal cancer tissue specimens in comparison with healthy tissue specimens.Methods:
A total of 210 tissue samples including 70 adenocarcinoma colorectal tissue, 70 adenomatous polyposis colorectal tissues, and 70 normal colorectal tissues were subjected to DNA extraction. The quality of the extracted DNA was confirmed by the amplification of a β-globin fragment using polymerase chain reaction (PCR). The presence of sod and glm genes were evaluated as Streptococcus gallolyticus and H. pylori presence markers by PCR method, respectively.Results:
Out of 210 subjects, 112 were male and the rest were female. The age of our patients ranged from 22 to 87 with an average of 54 years. None of the samples in two studied groups were positive for the sod and glm genes.Conclusions:
According to our results, S. gallolyticus subsp. gallolyticus and H. pylori might not be involved in colorectal cancer pathogenesis. More investigation on huge sample in different area might be clarified this results.Keywords
Colorectal Cancer Helicobacter pylori Streptococcus gallolyticus
1. Background
Worldwide, colorectal cancer is one of the main problems for healthcare systems, accounting for approximately one million of new cases yearly (1). Epidemiological studies have shown that colorectal cancer is the fourth most common cancer in Iran (2, 3). It has also been reported that the incidence of colorectal cancer in Iran is rapidly increasing (4). The development of malignancy is associated with several hereditary and environmental factors (2, 5, 6). Environmental factors, including infectious agents, have been estimated to be attributed for 20% of all cancers (1, 6).
The association between bacteria and colorectal cancer has been studied over four decades by using serological and molecular hints albeit the results were controversial (7). All these studies demonstrated that bowel bacterial infection is related to increased risk of colorectal cancer in certain individuals. These studies may also provide new aspects for more effective cancer prophylactic strategies and survival. Unlike stomach cancer development, which is mainly related to a single bacterial agent, various organisms including viruses, bacteria and parasitic agents have been postulated to contribute in colorectal cancer establishment (7). The tumorgenesis of bacteria might be attributed to overexpression of inflammatory cytokines as well as production of carcinogenic metabolites (8). Most of them induce chronic inflammation that will result in a competent procarcinogenic microenvironment (7, 9). The results of previous studies have revealed that the majority of colorectal cancers arise from adenomatous polyps (10). There are several bacteria that have been postulated as the causative agents of colorectal cancer including: Streptococcus gallolyticus subsp. gallolyticus (S. bovis biotype Ι) and Helicobacter pylori (7). The prevalence of H. pylori infection in the world population is around 50% (9). It has also been reported that 60% - 90% of Iranian population are infected by H. pylori (11).
H. pylori is a Gram-negative spiral shaped and motile bacterium that usually represents a major cause of gastric cancer, gastric lymphoma, gastric autoimmunity and peptic ulcer diseases (12). Induction of oxidative stress, carriage of cagA gene and DNA/RNA editing enzyme decreased apoptosis, and increased cell proliferation which are among factors responsible for H. pylori carcinogenesis (9, 12). Previous studies have revealed that H. pylori might slightly increase the risk of colorectal cancer (2, 13). H. pylori are also associated with an increased risk of pancreatic and esophageal/gastric cancer (14).
On the other hand, other bacterial agents that have been reported to be ambiguously associated with colorectal cancer is S. gallolyticus subsp. gallolyticus (7). As a gut commensal bacterium, it is thought to be resident in 2.5% - 15% of individuals (5). Moreover, infectious endocarditis from S. gallolyticus subsp. gallolyticus associated with colorectal cancer has been reported (15). It has been proposed that an increase in concentrations of cyclo-oxygenase2, interleukin 8 and cell proliferation might be involved in S. gallolyticus subsp. gallolyticus carcinogenesis (7).
Results from epidemiological studies are controversial, suggesting the association of colorectal cancer with many environmental factors such as high-fat diets and obesity. These findings maximally reveal the modest risk of these factors in colorectal cancer (16). Although over the past 30 years, direct evidences regarding the role of infectious causes in human cancer has been reported relevantly, but slight consideration has been given to the role of bacterial infection in the pathogenesis of colorectal cancer.
2. Objectives
Therefore, the aim of this study was to investigate the presence of S. gallolyticus subsp. gallolyticus and H. pylori in colorectal cancer tissue specimens in comparison with normal tissue in order to reveal the possible contribution of these bacteria in the etiology of colorectal cancer in Shiraz, south of Iran.
3. Methods
3.1. Study Population
From January 2012 to December 2013, 70 adenocarcinoma colorectal tissues, 70 adenomatous polyposis colorectal tissues, and 70 normal colorectal tissues as paraffin-embedded biopsy specimens were collected and included in this study. Samples were collected from Faghihi hospital, a major teaching hospital affiliated to Shiraz University of Medical Sciences. The study was approved by the medical ethics committee of the University and informed consent was obtained before sample collection.
3.2. Histopathology Examination
Colonoscopy and surgical biopsies of the normal colonic mucosa and colorectal cancer tissue were processed for molecular analysis and histopathology. Histological examination of Hematoxillin and Eosin-stained sections were performed by pathologists blinded to infection status using a standard Zeiss Axiophot microscope for the presence of malignant cells (Figure 1).
Photomicrograph of Human Colorectal Tissue
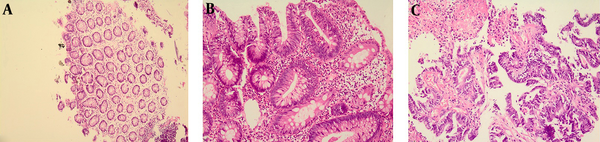
3.3. DNA Extraction
Seven sections (10 µm) of paraffin-embedded block were cut and placed in a 1.5 mL micro tube. A first step of de-paraffinization was performed by adding 1.2 mL of xylene to the 1.5 mL tubes containing the tissue section. After vortex and incubation of the tubes for 5 minutes at room temperature, the tubes underwent centrifugation at 14000 rpm for 5 minutes. Subsequently, the supernatant was removed and 1 mL of absolute ethanol was added to each tube and incubated for 5 minutes at room temperature. Finally, the tubes underwent centrifugation at 14000 rpm for 5 minutes and the supernatant was removed. Both steps were repeated once. In the next step, the tubes were incubated at 37°C on a heating block until the total evaporation of the ethanol. The DNA was then extracted using a QIAamp DNA minikit (Qiagen, Düsseldorf, Germany) according to the manufacturer’s instructions. The extracted DNA was stored at -20°C until testing.
3.4. PCR Analysis
All extracted DNA samples were initially subjected to PCR with consensus primers PCO3/PCO4 (β-globin) to verify the quality of the extracted DNA. PCR was performed in a total volume of 25 μL, containing 1 mM MgCl2 (CinnaGene, Iran), 200 μM (each) deoxyribonucleotide triphosphates solution (dNTPs) (CinnaGene, Iran), 1X reaction buffer (CinnaGene, Iran), 1U Taq DNA polymerase (CinnaGene, Iran), and 1 μM each specific primers (Table 1). PCR tests for β-globin were carried out as follows: 10 minutes initial denaturation at 94°C, 35 cycles of denaturation at 94°C for 45 seconds, annealing at 44°C for 45 seconds, extension at 72°C for 1 minute, and final extension at 72°C for 10 minutes. The detection of sod and glm genes were performed in samples which were positive for β-globin gene using specific primers (Table 1). The following PCR conditions were used for sod gene of S. gallolyticus subsp. gallolyticus: 5 minutes initial denaturation at 94°C, 50 cycles of denaturation at 94°C for 1 minute, annealing at 48°C for 30 seconds, extension at 72°C for 1 minute, and final extension at 72°C for 7 minutes. The PCR reactions using specific glmF/glmR primers were performed using the following steps: 5 minutes initial denaturation at 95°C, 50 cycles of denaturation at 95°C for 45 seconds, annealing at 50°C for 45 seconds, extension at 72°C for 35 seconds, and final extension at 72°C for 10 minutes. Then PCR products were loaded into 1.5% agarose gel and stained with 1% ethidium bromide and visualized under UV light (Figure 2).
To ensure reliability of test, positive controls including S. gallolyticus subsp. gallolyticus (ATCC 49475) and H. pylori (ATCC 26695) were also enrolled in each run.
The Sequences and Other Characteristics of Primers Used in This Study
| Locus | Primers | 5′ to 3′ Sequence | Size, bp |
|---|---|---|---|
| β-globin | PCO3 | 5′-ACACAACTGTGTTCACTAGC-3′ | 110 |
| PCO4 | 5′-CAACTTCATCCACGTTCACC-3′ | ||
| Sod | sod-F | 5′-CAATGACAATTCACCATGA-3′ | 408 |
| sod-R | 5′-TTGGTGCTTTTCCTTGTG-3′ | ||
| Glm | glm-F | 5′-AGCTTTTAGGGGTGTTAGGGGTTT-3′ | 294 |
| glm-R | 5′-AAGCTTACTTTCTAACACTAACGC-3′ |
Photographs of Gel Electrophoresis
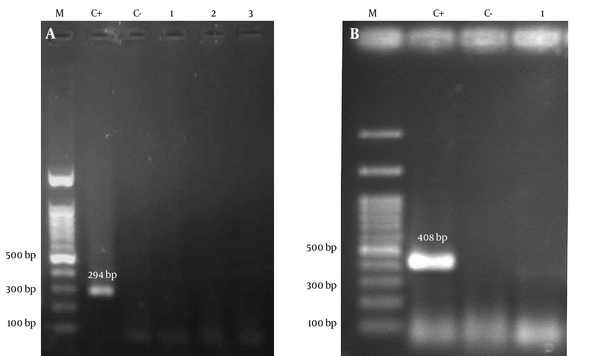
4. Results
A total of 210 patients participated in the study. There were 112 males (53.0%) and 98 females (47.0%). The patients’ age ranged between 22 and 87 years and the mean age was 54 years. Out of 140 subjects in the study group, 77 were male and 63 were female. Out of 70 cases in the control group, 35 were male and the rest were female. The age of most patients was between 40 and 60 years. The sub-site anatomic distribution of the tissues based on anatomic position was classified as colon (n = 89), rectum (n = 28), sigmoid (n = 24), and others (n = 69).
Molecular detection of H. pylori and S. gallolyticus subsp. gallolyticus revealed interesting finding. All of samples including 70 adenocarcinoma colorectal tissues, 70 adenomatous polyposis colorectal tissue and 70 normal colorectal specimens were negative for H. pylori and S. gallolyticus subsp. gallolyticus as revealed by no signal from sod and gml PCR amplifications.
5. Discussion
Colorectal cancer is one of the most common types of cancer worldwide (1). Sporadic occurrence and heterogenetic nature of colorectal cancer, including many factors, may be involved in colorectal cancer (7). It has been reported that infectious agents including bacteria might be associated with some malignancies such as colorectal cancer (1). During the past three decades, many studies have been performed to reveal the association between bacterial agents and this disease (13, 17). Among them, especially there are several studies that have shown the association between S. gallolyticus subsp. gallolyticus and H. pylori with colorectal cancer (13, 17). The heterogenetic nature of colorectal cancer has led to many epidemiological associations with causes of this disease. As our understanding about the molecular mechanism of colorectal cancer pathogenesis is growing fast, the microbe-epithelial interactions is a possible mechanism underlying tumor development (7).
Previously, we reported a low frequency of papillomavirus infection in colorectal cancer (18). In our new study, the study population was also evaluated for bacterial infection. Regarding PCR results, none of the colorectal tissues from patients and healthy subjects were positive for S. gallolyticus subsp. gallolyticus. The growth of S. gallolyticus in samples obtained from colorectal mucosa in a study performed by Potter et al. (19) did not show increased frequency in patients compared to the normal control group. In a study performed by Norfleet and Mitchell (20), the prevalence of S. gallolyticus infection was 3% (1 out of 33) in adenomas biopsies, 0% (0 out of 6) in adenocarcinomas, 0% (0 out of 14) in non-neoplastic polyps and 2.5% (1 out of 40) biopsies from normal mucosa. The results of both studies are inconsistent with our study which found no statistically significant association between S. gallolyticus infection and colorectal cancer. In contrast, Abdulamir et al. (17) reported a significant association between S. gallolyticus and colorectal cancer. Using molecular techniques, they showed a frequency of 48.7% (19 out of 39) for S. gallolyticus DNA sequences in colorectal tissue samples versus 4% (2 out of 50) in the normal mucosa of the control subjects.
In agreement with our results, Darjee et al. (21) reported no association between the seroprevalence of specific S. gallolyticus IgG in colorectal cancer patients and healthy control group. In a recent study from Malaysia, Al-Jashamy et al. (22) showed that 24.7% (41 of 166) of cancer cases were infected by S. gallolyticus. They also reported that in 48.6% (19 out of 41) of the stool specimens from the patients with colonic polyps, adenocarcinomas and inflammatory bowel diseases S. gallolyticus isolates were present. Moreover, Abdulamir et al. (23) showed higher levels of serum IgG antibodies against S. gallolyticus in the sera of colorectal cancer patients in comparison with the control subjects.
According to the above mentioned studies, the association between S. gallolyticus and colorectal cancer remains controversial. This controversy may arise from genetic background as well as geographical differences of studied cases from different areas. Besides, discrepancies may also be related to the different methods used for detection of S. gallolyticus, as well as the different specimens used in different studies. Indeed, S. bovis is one of the normal flora of human gastrointestinal tract.
In the present study, 70 adenocarcinoma colorectal tissues, 70 adenomatous polyposis colorectal tissues and 70 normal colorectal specimens were all negative for the presence of H. pylori glm gene sequence. In a study by Buhajic et al. (24), 1.2% and 6% of the tissue samples from patients with colorectal cancer and tissues from healthy control group were positive for H. pylori infection, respectively. Statistically, these results showed no correlation between H. pylori PCR positivity and colorectal cancer. In another study performed in Kashmir, 9.3% (8 out of 86) of cases were positive for H. pylori DNA sequences (25). Salehi et al. (2) reported the presence of glm gene sequence in 34.5% (20 of 58) of colorectal cancer tissues but not in normal tissues. In an investigation performed by Grahn et al. (26), 16S rDNA sequences of H. pylori was identified in 26% (11 out of 42) and 29% (10 out of 35) of colon and rectum cancer biopsies, respectively. The frequency of Helicobacter DNA sequences between the colon and rectum tumor biopsies were not significantly different.
Regarding the above-mentioned reports, the results of our study is consistent with those of studies performed by Buhajic et al. and Sameer et al. (24, 25) showing not a statistically significant association between the presence of H. pylori and colorectal cancer neoplasia. Although in studies conducted by Salehi et al. and Grahn et al. the association between H. pylori and colorectal cancer acclaimed to be significant, this association may be due to the type of samples (fresh versus paraffin-embedded samples). Furthermore, in two different sero-epidemiologic studies for investigation on H. pylori IgG prevalence and colorectal cancer, there was not any statistically significant association between H. pylori seropositivity and colorectal neoplasia (27, 28). These findings are consistent with our results which did not show a significant association between H. pylori exposure and colorectal cancer.
In contrast, in a large study performed by Zhang et al. in Germany, 46.1% (790 out of 1712) and 40.1% (669 out of 1669) of colorectal cancer cases and control subjects were positive for specific IgG against H.pylori. Also, in another study conducted by Zumkeller et al., 51% (195 of 384) of cancer patients and 44% (205 of 467) of matched control subjects were positive for antibodies to H. pylori and CagA. Both studies showed that this bacterium may be related to small but clinically relevant risk for development of colorectal cancer (29, 30). In a large cross-sectional study in South Korea, Hong et al. (13) showed that the prevalence of colorectal adenoma and advanced adenoma in the H.pylori positive group was significantly higher than in H. pylori negative group. They also showed that the presence of H. pylori is an important risk factor, but with limited importance in colorectal cancer. Although serology is the most common approach used to assess the association between H. pylori infection and colorectal cancer, it is to be noted that the increase in IgG antibody serum levels should be the results of colorectal H. pylori infection, not other sites such as stomach. Serologic assays do not identify the site of infection, so doing site-specific tests for evaluation of the etiologic relationship between infectious agents and colorectal cancer is necessary. This issue supports the difference in our findings versus others, done by serological methods. Such controversy may also arise from the differences in genetic and geographical background.
In conclusion, our study does not support the association between S. gallolyticus and H. pylori infections with colorectal cancer. More studies with more cases in different areas are needed to clarify the association between S. gallolyticus and H. pylori infection and colorectal cancer.
Acknowledgements
References
-
1.
Antonic V, Stojadinovic A, Kester KE, Weina PJ, Brucher BL, Protic M, et al. Significance of infectious agents in colorectal cancer development. J Cancer. 2013;4(3):227-40. [PubMed ID: 23459622]. https://doi.org/10.7150/jca.5835.
-
2.
Salehi Z, Miri M, Aminian K, Mansour-Ghanaei F. Helicobacter pylori infection and colorectal cancer in Guilan province of Iran. Ann Biol Res. 2011;2(1):32-9.
-
3.
Sadjadi A, Malekzadeh R, Derakhshan MH, Sepehr A, Nouraie M, Sotoudeh M, et al. Cancer occurrence in Ardabil: results of a population-based cancer registry from Iran. Int J Cancer. 2003;107(1):113-8. [PubMed ID: 12925965]. https://doi.org/10.1002/ijc.11359.
-
4.
Azadeh S, Moghimi-Dehkordi B, Fatem SR, Pourhoseingholi MA, Ghiasi S, Zali MR. Colorectal cancer in Iran: an epidemiological study. Asian Pac J Cancer Prev. 2008;9(1):123-6. [PubMed ID: 18439090].
-
5.
Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agent Cancer. 2013;8(1):11. [PubMed ID: 23537317]. https://doi.org/10.1186/1750-9378-8-11.
-
6.
Sarvari J, Mojtahedi Z, Kuramitsu Y, Fattahi MR, Ghaderi A, Nakamura K, et al. Comparative Proteomics of Sera From HCC Patients With Different Origins. Hepat Mon. 2014;14(1):13103. [PubMed ID: 24497876]. https://doi.org/10.5812/hepatmon.13103.
-
7.
Collins D, Hogan AM, Winter DC. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2011;12(5):504-12. [PubMed ID: 21067973]. https://doi.org/10.1016/S1470-2045(10)70186-8.
-
8.
Mantzaris GJ. Helicobacter pylori and colorectal cancer: Is there any link? Ann Gastroenterol. 2004;17(1):17-8.
-
9.
Nishizawa T, Suzuki H. Gastric Carcinogenesis and Underlying Molecular Mechanisms: Helicobacter pylori and Novel Targeted Therapy. Biomed Res Int. 2015;2015:794378. [PubMed ID: 25945346]. https://doi.org/10.1155/2015/794378.
-
10.
Atkin WS, Morson BC, Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N Engl J Med. 1992;326(10):658-62. [PubMed ID: 1736104]. https://doi.org/10.1056/NEJM199203053261002.
-
11.
Hosseini E, Poursina F, de Wiele TV, Safaei HG, Adibi P. Helicobacter pylori in Iran: A systematic review on the association of genotypes and gastroduodenal diseases. J Res Med Sci. 2012;17(3):280-92. [PubMed ID: 23267382].
-
12.
Bazargani A, Khoramrooz SS, Kamali-Sarvestani E, Taghavi SA, Saberifiroozi M. Association between peroxisome proliferator-activated receptor-gamma gene polymorphism (Pro12Ala) and Helicobacter pylori infection in gastric carcinogenesis. Scand J Gastroenterol. 2010;45(10):1162-7. [PubMed ID: 20568969]. https://doi.org/10.3109/00365521.2010.499959.
-
13.
Hong SN, Lee SM, Kim JH, Lee TY, Kim JH, Choe WH, et al. Helicobacter pylori infection increases the risk of colorectal adenomas: cross-sectional study and meta-analysis. Dig Dis Sci. 2012;57(8):2184-94. [PubMed ID: 22669208]. https://doi.org/10.1007/s10620-012-2245-x.
-
14.
Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784-9. [PubMed ID: 11556297]. https://doi.org/10.1056/NEJMoa001999.
-
15.
Waisberg JMC, Pimenta J. Infectious endocarditis from Streptococcus bovis associated with colonic carcinoma: Case report and literature review. Arq Gastroenterol. 2002;39(3):177-80. https://doi.org/10.1590/S0004-28032002000300008.
-
16.
Alexander DD, Cushing CA, Lowe KA, Sceurman B, Roberts MA. Meta-analysis of animal fat or animal protein intake and colorectal cancer. Am J Clin Nutr. 2009;89(5):1402-9. [PubMed ID: 19261724]. https://doi.org/10.3945/ajcn.2008.26838.
-
17.
Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249. [PubMed ID: 20846456]. https://doi.org/10.1186/1476-4598-9-249.
-
18.
Mahmoudvand S, Safaei A, Erfani N, Sarvari J. Presence of Human Papillomavirus DNA in Colorectal Cancer Tissues in Shiraz, Southwest Iran. Asian Pac J Cancer Prev. 2015;16(17):7883-7. [PubMed ID: 26625815]. https://doi.org/10.7314/APJCP.2015.16.17.7883.
-
19.
Potter MA, Cunliffe NA, Smith M, Miles RS, Flapan AD, Dunlop MG. A prospective controlled study of the association of Streptococcus bovis with colorectal carcinoma. J Clin Pathol. 1998;51(6):473-4. [PubMed ID: 9771449].
-
20.
Norfleet RG, Mitchell PD. Streptococcus bovis does not selectively colonize colorectal cancer and polyps. J Clin Gastroenterol. 1993;17(1):25-8. [PubMed ID: 8409294]. https://doi.org/10.1097/00004836-199307000-00008.
-
21.
Darjee R, Gibb AP. Serological investigation into the association between Streptococcus bovis and colonic cancer. J Clin Pathol. 1993;46(12):1116-9. [PubMed ID: 8282836]. https://doi.org/10.1136/jcp.46.12.1116.
-
22.
Al-Jashamy K, Murad A, Zeehaida M, Rohaini M, Hasnan J. Prevalence of colorectal cancer associated with Streptococcus bovis among inflammatory bowel and chronic gastrointestinal tract disease patients. Asian Pac J Cancer Prev. 2010;11(6):1765-8. [PubMed ID: 21338230].
-
23.
Abdulamir AS, Hafidh RR, Mahdi LK, Al-jeboori T, Abubaker F. Investigation into the controversial association of Streptococcus gallolyticus with colorectal cancer and adenoma. BMC Cancer. 2009;9:403. [PubMed ID: 19925668]. https://doi.org/10.1186/1471-2407-9-403.
-
24.
Bulajic M, Stimec B, Jesenofsky R, Kecmanovic D, Ceranic M, Kostic N, et al. Helicobacter pylori in colorectal carcinoma tissue. Cancer Epidemiol Biomarkers Prev. 2007;16(3):631-3. [PubMed ID: 17372266]. https://doi.org/10.1158/1055-9965.EPI-06-1031.
-
25.
Sameer AS, Abdullah S, Nissar S, Rasool R, Mudassir Baba S, Siddiqi MA. Helicobacter pylori in colorectal neoplasms of Kashmiri patients: What is the prevalence? Clin Cancer Investig J. 2012;1(1):23-5. https://doi.org/10.4103/2278-0513.95015.
-
26.
Grahn N, Hmani-Aifa M, Fransen K, Soderkvist P, Monstein HJ. Molecular identification of Helicobacter DNA present in human colorectal adenocarcinomas by 16S rDNA PCR amplification and pyrosequencing analysis. J Med Microbiol. 2005;54(Pt 11):1031-5. [PubMed ID: 16192433]. https://doi.org/10.1099/jmm.0.46122-0.
-
27.
Fireman Z, Trost L, Kopelman Y, Segal A, Sternberg A. Helicobacter pylori: seroprevalence and colorectal cancer. Isr Med Assoc J. 2000;2(1):6-9. [PubMed ID: 10892362].
-
28.
Siddheshwar RK, Muhammad KB, Gray JC, Kelly SB. Seroprevalence of Helicobacter pylori in patients with colorectal polyps and colorectal carcinoma. Am J Gastroenterol. 2001;96(1):84-8. [PubMed ID: 11197293]. https://doi.org/10.1111/j.1572-0241.2001.03355.x.
-
29.
Zhang Y, Hoffmeister M, Weck MN, Chang-Claude J, Brenner H. Helicobacter pylori infection and colorectal cancer risk: evidence from a large population-based case-control study in Germany. Am J Epidemiol. 2012;175(5):441-50. [PubMed ID: 22294430]. https://doi.org/10.1093/aje/kwr331.
-
30.
Zumkeller N, Brenner H, Chang-Claude J, Hoffmeister M, Nieters A, Rothenbacher D. Helicobacter pylori infection, interleukin-1 gene polymorphisms and the risk of colorectal cancer: evidence from a case-control study in Germany. Eur J Cancer. 2007;43(8):1283-9. [PubMed ID: 17446060]. https://doi.org/10.1016/j.ejca.2007.03.005.