Abstract
Introduction:
Campylobacter jejuni (C. jejuni) and Campylobacter coli (C. coli) are the most common campylobacter species related to human gastroenteritis. Due to their large similarity, these two species are not differentiated in laboratories. In this report, the coinfection with C. jejuni and C. coli was studied in two pediatric patients. The aim of the present report was to determine if simultaneous coinfection with C. jejuni and C. coli, with different antibiotic profiles, could happen.Case Presentation:
In the present report, two patients clinically diagnosed with bacillary dysentery, showing fever and pus in their stool and undergoing treatment with cotrimoxazole, were microbiologically investigated through the modified Gram stain, culture and duplex PCR for diagnosing C. jejuni and C. coli. Based on microbiological and molecular results, coinfection with C. jejuni and C. coli were determined in both patients. Campylobacters isolated from patients were resistant to erythromycin, tetracycline, ciprofloxacin, ampicillin, gentamicin, and nalidixic acid. Also, in both patients, C. jejuni was sensitive to cotrimoxazole and ceftriaxone. In contrast, isolated C. coli were resistant to cotrimoxazole and sensitive to ceftriaxone.Conclusions:
The two patients were simultaneously infected with C. jejuni and C. coli and both were carried all the antibiotic resistant genes under study. In spite of the sensitivity of C. jejuni to cotrimoxazole, no improvement was observed for C. coli due to its resistance to this antibiotic. This finding emphasizes on the important role of microbiology investigation once empirical therapy is needed. This issue must be taken seriously in pediatric hospitals.Keywords
Pediatric Coinfection Campylobacter jejuni Campylobacter coli Dysentery Iran
1. Introduction
World Health Organization (WHO) has considered diarrhea, after AIDS, as the second leading cause of death among children under 5. This means the death of around 760000 children every year. Campylobacter is one of the major factors affecting about 400 million people worldwide each year (1). The infections of the genus campylobacter have been reported as the most common cause of acute diarrhea, especially among children under 3 and the elderly (2). C. jejuni and C. coli are the most common campylobacter species related to human gastroenteritis, and their frequency is 3 - 4 times higher than that of the other bacterial enteropathogens like Salmonella and pathogenic Escherichia coli in patients with gastrointestinal infections (1). Coinfection is the concurrent infection of a host by multiple pathogenic species (3). In Iran, campylobacter has rarely been reported as a causative agent of gastroenteritis. Given that C. jejuni is often the causative agents of gastroenteritis and that C. coli causes 2% - 5% of infections in USA, the microbiological diagnoses are usually based on the identification of C. jejuni (4).
In this report, out of the most common bacterial factors developing dysentery, the coinfection with C. jejuni and C. coli was investigated in two patients clinically through laboratory tests.
2. Case Presentation
2.1. The First Patient
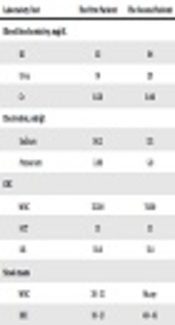
The first patient was a 12-month-old infant boy who was the third child of the family, weighed 9.1 kg, and lived in Arak. He was never hospitalized, had performed vaccination according to the national protocol. Also, he had undergone empirical therapy with cotrimoxazole and acetaminophen for 7 days due to his diarrhea and was clinically diagnosed with shigellosis; however, he had not shown any sign of improvement. Moreover, he had a 38°C fever, loose bloody diarrhea, abdominal cramps and mild dehydration without vomiting for two days before being referred to Amir Kabir Hospital in Arak which led to his hospitalization in isolation ward. Table 1 shows the results of the laboratory tests for this patient.
The Results of the Laboratory Tests for the Patient 1 and the Patient 2
| Laboratory Test | The First Patient | The Second Patient |
|---|---|---|
| Blood biochemistry, mg/dL | ||
| BS | 93 | 94 |
| Urea | 19 | 20 |
| Cr | 0.50 | 0.48 |
| Electrolyte, mEq/L | ||
| Sodium | 141.3 | 135 |
| Potassium | 3.88 | 5.0 |
| CBC | ||
| WBC | 11500 | 7600 |
| HCT | 35 | 35 |
| HB | 10.8 | 11.8 |
| Stool exam | ||
| WBC | 30 - 35 | Many |
| RBC | 10 - 12 | 40 - 45 |
The patient was investigated for campylobacter as the routine stool culture tests were negative for Salmonella and Shigella. According to the patient’s parents, ten days ago the child had tasted a piece of unwashed chicken several times while his mother was washing them. The sample was negative for Salmonella, Shigella, and pathogenic E. coli (both phenotypically and genotypically). However, the campylobacter tests were carried out using the modified Gram stain with the carbol fuchsin being heated for 5 minutes, (Modified charcoal cefoperazone deoxycholate agar) mCCDA medium (Ibresco, Iran), passing through a 0.45nm paper filter (Sartorius, Germany) and brucella agar medium (Merck, Germany) with 10% defibrinated sheep blood. Also, the oxidase and catalase differential tests and the sodium hippurate hydrolysis test (Merck, Germany) were carried out on the suspected colonies and all the results were positive (5).
After extracting the DNA from the stool sample and performing the Duplex PCR with specific genes being mapA to identify C. jejuni and ceuE, and C. coli, the results for both genes were positive (6). Also, the results of sequencing with the products of PCR from both genes and the blast verified the simultaneous existence of C. jejuni and C. coli. Two reference strains, ATCC 33560 (Campylobacter jejuni) and ATCC 33559 (Campylobacter coli), from the Department of Microbiology, Faculty of Medical Sciences, Arak University of Medical Sciences were used as the positive controls for the analysis of the fecal sample isolates.
According to the CLSI 2016 method (7), an antibiogram was performed on the isolated campylobacter colonies. It was found that the antibiotic was resistant to erythromycin, tetracycline, ciprofloxacin, ampicillin, gentamicin, and nalidixic acid and the bacteria were sensitive to ceftriaxone (Mast Diagnostics, UK). DNA extraction was performed directly from the stool samples and C. jejuni isolates using QIAamp DNA Stool Minikit (Qiagen GmbH, Hilden, Germany) according to the protocol. Also, the PCR was genotypically carried out from the antibiotic resistant genes tet (O), Oxa61, CmeB, Sul1, sul2, qac, gyrA4, gyrA5, gyrA6, and qnrS, and all the results were positive. In addition, the existence of mutation was confirmed by investigating the nucleotide sequence of 23srRNA, proliferated with specific primers (6, 8-10). From each positive gene, one sample was used for sequencing of the PCR product of isolates by Gene Fanavaran Company and the sequence was identified by BLAST analysis.
After hospitalization, the empirical therapy started with prescribing a half-liter 1.3 - 2.3 infusion, half-liter saline solution for intravenous infusion, and ceftriaxone 500 mg (Rocephin).
The symptoms of the patient disappeared three days after the onset of treatment, and then the patient was discharged.
2.2. The Second Patient
The other patient was a 5-month-old infant boy living in Arak. He was the first child of the family with no history of hospitalization and surgery. It was said that he had experienced symptoms like those of cold (fever, cough, sweating) for four days and was consequently referred to a physician. He was under treatment for cold with cetirizine and cotrimoxazole but did not show any sign of improvement. Also, he had fever, bloody diarrhea, and lots of white and red blood cells in his stool 2 days before being referred to Amir Kabir Hospital. The results of the laboratory tests for the patient are shown in Table 1. An abdominal ultrasonography was requested for the patient during his stay and the result was normal. Also, like the first patient, the laboratory tests were taken for the second patient as well (both phenotypically and genotypically).
Like the first patient, the results were phenotypically and genotypically positive for C. jejuni and C. coli for this patient. The patient received ceftriaxone 500 mg. Being recovered, he was discharged after three days.
3. Discussion
In this report, two patients were identified to have coinfection with C. jejuni and C. coli. Commonly, simultaneous detection of both species of C. jejuni and C. coli is unusual in clinical laboratories. The patient’s manifestations were diarrhea followed by abdominal pain and fever, as well as the appearance of blood and mucus in the stool. Therefore, it can be concluded that these clinical symptoms can be considered as the clinical indexes for the diagnosis of campylobacteriosis in the campylobacter infections. The present report shows that fever, diarrhea, leukocytes, and blood in the stool are the most common clinical symptoms of coinfection with two species of C. jejuni and C. coli. Therefore, in clinical diagnosis, there is no distinction between the clinical manifestations of coinfection with two campylobacter species.
Previously, four cases of coinfection with C. jejuni and C. coli were observed in England in 2001, and 3.6% coinfection with different campylobacter strains has been reported in 1991. Also, 1.8% coinfection with C. jejuni and C. coli was observed in Brazil and India in 2010, and 22 children were reported to have coinfection with other enteropathogenic species of E. coli, rotavirus, and Salmonella in Poland in 2013 (2, 11-13).
In this report, the resistance of the diagnosed organism in the two patients was investigated phenotypically and genotypically and it was revealed that they were carrying several antibiotic resistant genes. In a study by Feizabadi et al. in Tehran, Iran, the amount of resistance to antimicrobial agents were as follows: ciprofloxacin (61.7%), ceftazidime (47%), carbenicillin (35%), tetracycline (20.5%), cefotaxime (14.7%), ampicillin (11.7%), neomycin erythromycin and chloramphenicol (2.9%), gentamicin, streptomycin, imipenem, and colistin (0.0%) (14). Considering the high resistance of C. coli to erythromycin, which is the drug of choice for treating C. jejuni (15), the evidence of the present report shows that if one is coinfected with C. jejuni and C. coli, their antibiotic resistance should be taken into consideration. It might also require a different antibiotic treatment.
In this report, the coinfection with C. jejuni and C. coli in one individual was reported for the first time in Iran. The coinfection with C. jejuni and C. coli in one individual is less addressed in other parts of the world.
This is the first report investigating the clinical symptoms of coinfection with two campylobacter species as no other report has been issued, up to now, on investigating the clinical symptoms of coinfection with two campylobacter species.
Indeed, it is not true that only one organism can always be the cause of infection, but it is recommended that all the enteropathogenic organisms should be considered in the stool sample culture, because ignoring the coinfection with multiple organisms can lead to a wrong or incomplete treatment as well as the longer stay of the patient in the hospital. The present report attempts to push laboratories to focus on the coinfection since the possibility of simultaneous existence of more than one factor causing infection can always happen.
Acknowledgements
References
-
1.
Hailemariam S. Prevalence, associated risk factors and antimicrobial susceptibility pattern of thermophilic Campylobacter spp. of ovine carcass at Addis Ababa Abattoir Enterprise, Ethiopia [dissertation]. Addis Ababa, Ethiopia: Addis Ababa University; 2014.
-
2.
Grzybowska-Chlebowczyk U, Kalita B, Flak-Wancerz A, Jasielska M, Więcek S, Wojcieszyn M, et al. Clinical course of Campylobacter infections in children. Pediatria Polska. 2013;88(4):329-34. https://doi.org/10.1016/j.pepo.2013.05.004.
-
3.
Diuk-Wasser MA, Vannier E, Krause PJ. Coinfection by ixodes tick-borne pathogens: Ecological, epidemiological, and clinical consequences. Trends Parasitol. 2016;32(1):30-42. [PubMed ID: 26613664]. [PubMed Central ID: PMC4713283]. https://doi.org/10.1016/j.pt.2015.09.008.
-
4.
Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology. Philadelphia: Elsevier Health Sciences; 2015.
-
5.
Garcia LS. Clinical microbiology procedures handbook. American Society for Microbiology Press; 2010.
-
6.
Tsang RS, Figueroa G, Bryden L, Ng L. Flagella as a potential marker for Campylobacter jejuni strains associated with Guillain-Barre syndrome. J Clin Microbiol. 2001;39(2):762-4. [PubMed ID: 11158146]. [PubMed Central ID: PMC87815]. https://doi.org/10.1128/JCM.39.2.762-764.2001.
-
7.
Patel JB. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute; 2017.
-
8.
Obeng AS, Rickard H, Sexton M, Pang Y, Peng H, Barton M. Antimicrobial susceptibilities and resistance genes in Campylobacter strains isolated from poultry and pigs in Australia. J Appl Microbiol. 2012;113(2):294-307. [PubMed ID: 22672511]. https://doi.org/10.1111/j.1365-2672.2012.05354.x.
-
9.
Zirnstein G, Li Y, Swaminathan B, Angulo F. Ciprofloxacin resistance in Campylobacter jejuni isolates: Detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J Clin Microbiol. 1999;37(10):3276-80. [PubMed ID: 10488192]. [PubMed Central ID: PMC85547].
-
10.
Hopkins KL, Wootton L, Day MR, Threlfall EJ. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J Antimicrob Chemother. 2007;59(6):1071-5. [PubMed ID: 17434878]. https://doi.org/10.1093/jac/dkm081.
-
11.
da Silva Quetz J, Lima IF, Havt A, de Carvalho EB, Lima NL, Soares AM, et al. Campylobacter jejuni and Campylobacter coli in children from communities in Northeastern Brazil: Molecular detection and relation to nutritional status. Diagn Microbiol Infect Dis. 2010;67(3):220-7. [PubMed ID: 20542202]. [PubMed Central ID: PMC2886016]. https://doi.org/10.1016/j.diagmicrobio.2010.02.025.
-
12.
Nichols GL, Richardson JF, Sheppard SK, Lane C, Sarran C. Campylobacter epidemiology: A descriptive study reviewing 1 million cases in England and Wales between 1989 and 2011. BMJ Open. 2012;2(4). [PubMed ID: 22798256]. [PubMed Central ID: PMC3400078]. https://doi.org/10.1136/bmjopen-2012-001179.
-
13.
Ghosh R. Increasing antimicrobial resistance of campylobacter jejuni isolated from paediatric diarrhea cases in a tertiary care hospital of New Delhi, India. Journal of Clinical and Diagnostic Research. 2013. https://doi.org/10.7860/jcdr/2013/5267.2738.
-
14.
Feizabadi MM, Dolatabadi S, Zali MR. Isolation and drug-resistant patterns of Campylobacter strains cultured from diarrheic children in Tehran. Jpn J Infect Dis. 2007;60(4):217-9. [PubMed ID: 17642538].
-
15.
Luangtongkum T, Jeon B, Han J, Plummer P, Logue CM, Zhang Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009;4(2):189-200. [PubMed ID: 19257846]. [PubMed Central ID: PMC2691575]. https://doi.org/10.2217/17460913.4.2.189.