Abstract
Background:
The incidence of coagulase-negative staphylococci (CNS) isolates, as causes of hospital-acquired infections, is increasing annually and it is a truly global challenge.Objectives:
The current study aimed at assessing aminoglycoside resistance genes among CNS isolated from hospitalized patients and investigating the prevalence of methicillin and aminoglycoside resistance genes using molecular methods.Methods:
A total of 103 species of coagulase negative staphylococci were isolated from clinical specimens from August 2013 to November 2014. All the specimens were identified using conventional microbiological methods including colony morphology, Gram staining, catalase, and slide and tube coagulase. The isolates were subjected to the API-20 Staph identification kit. Antibiotic susceptibility tests were performed using Kirby-Bauer disc diffusion method. The methicillin and aminoglycoside resistant genes among coagulase negative staphylococci were detected by polymerase chain reaction (PCR) and sequencing methods.Results:
The most frequent clinically isolated coagulase negative staphylococci species were Staphylococcus hominis and S. haemolyticus (51.9%), S. epidermidis (21.7%), S. lugdunensis (9.8%), S. intermedius (4.9%), S. saprophyticus and S. simulans (3.9%), S. warneri (2.9%), as well as S. chromogenes, S. sciuri, S. caprae, S. schleiferi, and S. xylosus (0.98%). The overall rate of methicillin resistance among CNS species in the present study was 89 (86.4%). The resistance of CNS isolates against tested antibiotics was as follows: 74 (71.8%) to cefoxitin, 54 (52.4%) to kanamycin, 51 (49.5%) to gentamicin, 45 (43.7%) to tobramycin, and 30 (29.1%) to amikacin. The prevalence of aminoglycoside resistance genes such as ant (4')-Ia, aac (6')/aph (2") and aph (3')-IIIa were 89 (86.4%), 87(84.5%), and 68(66%), respectively.Conclusions:
The presence of antibiotic resistance among coagulase negative Staphylococcus species, which cause nosocomial infections, has increased. Therefore, identification of coagulase negative staphylococci within 24 hours after development of enzymatic tests would be very useful in any clinical microbiology laboratory for effective treatment. Resistance to antibiotics, including aminoglycosides, develops quickly in CNS species where these antimicrobial agents are widely used. Thus, a better understanding of antibiotic resistance of CNS species is essential to eliminate antibiotic resistance in hospitals and society.Keywords
Coagulase Negative Staphylococcus API Test Aminoglycoside Resistance Genes
1. Background
Coagulase-negative staphylococci (CNS) infections are commonly associated with medical-device related infections, such as heart valves, pacemaker lines and boxes, cerebrospinal shunts, prosthetic joints, and also in immune-compromised patients, such as the patients with cancer and the ones undergoing chemotherapy and dialysis (1, 2).
Meanwhile, aminoglycoside antibiotics are one of the most used antibiotics in clinical practice that play an important role in the treatment of patients. Resistance to aminoglycosides develops quickly in CNS species among different clinical settings (3).
Inactivating the drug could be caused by aminoglycoside-modifying enzymes (AMEs) encoded within mobile genetic elements (4). Modifying enzymes are encoded with several distinct genes including aminoglycoside-6’-N-acetyltransferase/2”-O-phosphoryltransferase [aac(6’)/aph(2’’)], aminoglycoside-4’-O-nucleotidyltransferase I [ant(4’)-I], and aminoglycoside-3’-O-phosphoryltransferase III [aph(3’)-III] (5, 6). Subclasses are defined by the position of the catalytic sites, at a hydroxyl group (aph and ant) or at an -NH2 group (aac) (7). The ant(4’)-I mediates resistance to neomycin, kanamycin, tobramycin, and amikacin in staphylococci; while, aph(3’)-III can mediate resistance to neomycin and kanamycin (7). The distribution of AME genes largely depends on bacterial species, but the combination of AMEs is correlated with the use of aminoglycosides in specific geographic regions (8).
2. Objectives
The current study aimed at describing the identification of isolated CNS and investigating the presence of genes encoding AMEs to the aminoglycoside susceptibility pattern and methicillin resistance.
3. Methods
3.1. Bacterial Strains
From August 2013 to November 2014, a total of 103 CNS isolates were collected from clinical specimens of the patients admitted to 6 major hospitals located in different regions of Tehran, Iran. The isolates were identified by their phenotypic characteristics such as colony morphology, Gram staining, catalase, and slide and tube coagulase test results.
3.2. Identification and Susceptibility Testing
The identification of CNS species was carried out using API-Staph system. Each isolate was grown overnight on Muller-Hinton agar (Merck, Darmstadt, Germany) and added in an aliquot of the API-Staph medium to each tube of the gallery, enclosed within rigid plastic tray (API bioMerieux, France). The galleries were incubated for 24 hours at 37°C. The tested biochemical reactions included the fermentations of glucose, fructose, mannose, maltose, lactose, trehalose, mannitol, xylitol, melibiose, raffinose, xylose, sucrose, a-methyl glucoside, and N-acetyl glucosamine, the reduction of nitrate, the hydrolysis of arginine and urea, phosphatase activity, and the formation of acetoin from pyruvate. At the end of the incubation period, to demonstrate alkaline phosphatase activity, acetyl-methyl carbinol formation, and nitrate reduction, active reagents, containing β- naphthyl phosphate, sodium pyruvate, and potassium nitrate were added to the cupules.
3.3. Antibiotic Susceptibility Testing
All the isolates were tested, using a disc diffusion method, to evaluate resistance to gentamicin (GEN: 10 μg), tobramycin (TOB: 10 μg), kanamycin (KAN: 30 μg), amikacin (Ami: 30 μg), and cefoxitin (CEF: 30 μg) (MAST, Merseyside. U.K)according to the clinical and laboratory standards institute (CLSI) guidelines (9).
3.4. DNA Extraction and Amplification
DNA was extracted by a kit (Qiagen, Hilden, Germany). The polymerase chain reaction (PCR) was carried out in Intellectica thermo cycler (Techne, UK) by the PCR Master Kit (Cinna Clone Inc., Iran), according to the manufacturer’s guidelines. Primer sequences were used to detect mecA, ant (4΄)-Ia, aac (6΄)-Ie/aph (2˝), and aph (3΄)-IIIa genes; the fragment sizes are presented in Table 1. PCR condition was as follows: Initial denaturation at 95°C for 5 minutes followed by 30 cycles of denaturation at 95°C for 1 minute, annealing for mecA gene 1 minute at 57°C, aac (6’)/aph (2’’) 45 second at 56°C, ant (4’)-I 30 second at 48°C, and aph (3’)-III 45 second at 57.5°C, extension at 72°C for 30 second. Staphylococcus aureus USS1504, S. aureus BM3002/ piP52, and Enterococcus faecalis BM6217 were used as the positive controls for mecA, ant (4′)-Ia, aph (3′)-IIIa, aac (6′)/aph (2′′), respectively.
PCR Primers Used to Detect Aminoglycoside Resistance Genes
The final extension step was continued for another mecA gene for 8 minutes at 72°C and the rest of aminoglycoside gene for 60 seconds at 72°C. DNA fragments were analyzed using electrophoresis in a 1% agarose gel at 95 V for 45 minutes in 1 X Tris-Borate-ethylenediaminetetraacetic acid (TBE) containing ethidium bromide. Finally, sequencing of forward strand was performed by the Bioneer Company (Korea). The nucleotide sequences were analyzed using Chromas 1.45 and MEGA-4 software and BLAST at NCBI.
3.5. Statistical Analysis
Statistical analysis was performed using SPSS for Windows (version 17.0) (SPSS Inc., Chicago, IL) running Chi-squared test.
4. Results
A total of 103 strains of CNS (blood = 27, wound = 37, pus = 1, urine = 15, central nervous system (CNS) = 2, catheter = 4, sputum = 9, bronchoalveolar lavage fluid = 2, prosthetic joints = 1, and body fluids = 5) were recovered from 6 hospitals in Tehran, Iran.
From a total of 103 strains, 102 were identified using the API test; only one isolate, obtained from a prosthetic joint, could not be identified. Therefore, 102 strains were analyzed. Table 2 presents the identification list of the most likely species among hospitalized patients in Iran.
Frequency of CNS Species Identified by Commercial Kits
| Organisms | No. of Isolates (%) | No. of Isolates From | |||
|---|---|---|---|---|---|
| Wound Infection | Endocarditis and Blood Cultures | Urinary Tract Infections | Miscellaneous Infections | ||
| Staphylococcus hominis and Staphylococcus haemolyticus | 53 (51.9) | 21 | 5 | 11 | 16 |
| Staphylococcus epidermidis | 22 (21.7) | 5 | 15 | 1 | 1 |
| Staphylococcus lugdunensis | 10 (9.8) | 5 | 2 | 2 | 1 |
| Staphylococcus intermedius | 5 (4.9) | 2 | 2 | 1 | |
| Staphylococcus saprophyticus and Staphylococcus simulans | 4 (3.9) | 1 | 2 | 0 | 1 |
| Staphylococcus warneri | 3 (2.9) | 1 | 1 | 0 | 1 |
| Staphylococcus chromogenes | 1 (0.98) | 0 | 0 | 1 | 0 |
| Staphylococcus sciuri | 1 (0.98) | 0 | 0 | 0 | 1 |
| Staphylococcus caprae | 1 (0.98) | 1 | 0 | 0 | 0 |
| Staphylococcus schleiferi | 1 (0.98) | 1 | 0 | 0 | 0 |
| Staphylococcus xylosus | 1 (0.98) | 0 | 0 | 0 | 1 |
| Total identified no. (%) | 102 (100.0) | 37 | 27 | 15 | 23 |
In vitro susceptibility of the CNS isolates were tested to 5 antibiotics. A total of 54 isolates (52.4%) were resistant to kanamycin. Some strains of CNS were detected with intermediate resistance to kanamycin 4 (3.9%), tobramycin, and amikacin 2 (1.9%) (Table 3).
| Hospital | Antibiotics | ||||
|---|---|---|---|---|---|
| Tobramycin | Kanamycin | Amikacin | Gentamicin | Cefoxitin | |
| K | 17 (65.4) | 20 (76.9) | 20 (76.9) | 20 (76.9) | 25 (96.2) |
| M | 7 (30.4) | 7 (30.4) | 1 (4.3) | 8 (34.8) | 12 (52.2) |
| P | 8 (33.3) | 9 (37.5) | 4 (16.7) | 8 (33.3) | 18 (75) |
| MO | 6 (46.2) | 8 (61.5) | 2 (15.4) | 7 (53.8) | 9 (69.2) |
| S | 1 (11.1) | 3 (33.3) | 0 | 2 (22.2) | 3 (33.3) |
| T | 6 (75) | 7 (87.5) | 3 (37.5) | 6 (75) | 7 (87.5) |
| Total | 43.7 | 52.4 | 29.1 | 49.5 | 71.8 |
According to the results of the present study, the rate of resistance to kanamycin, gentamicin, tobramycin, and amikacin were 52.4%, 49.5%, 43.7% and 29.1%, respectively. Also, with regard to Table 4, the proportion of methicillin-resistant CNS (MRCNS), which showed resistance to the aminoglycosides in the present study, was higher than that of the methicillin-sensitive CNS (MSCNS) isolates (Figure 1).
The Distribution of Aminoglycoside Resistance Genes in Isolates of CNS in Relation to Methicillin Resistance
| Resistant Gene | Methicillin Sensitive CNS (n = 14) | Methicillin Resistance CNS (n = 89) |
|---|---|---|
| aac(6’)/aph(2’’) | 13 (92.9) | 74 (83.1) |
| aph(3’)-III | 9 (64.3) | 59 (66.3) |
| ant(4’)-I | 11 (78.9) | 78 (87.6) |
Detection of Genes Encoding mecA Gene in CNS Isolates by PCR

The highest prevalence of aminoglycoside resistance genes among CNS isolates was ant (4’)-I occurring in 87.6% of MRCNS species (Figure 2). However, the rate of aac (6’)/aph (2’’) was very high (Figure 3). The least common gene was aph (3’)-III, found in 66.3% of MRCNS species (Figure 4). The rate of coexistence of aac (6’)-Ie-aph (2’’) with aph (3’)-IIIa, and aac (6’)-Ie-aph (2’’) with ant (4’)-Ia was 65 (63%) and 77 (74%), respectively. The rate of coexistence of 3 AME genes was 57 (55.3%).
Detection of Genes Encoding ant (4’)-I Gene in CNS Isolated by PCR

Detection of Genes Encoding aac(6’)-aph(2’’) Gene in CNS Isolates by PCR
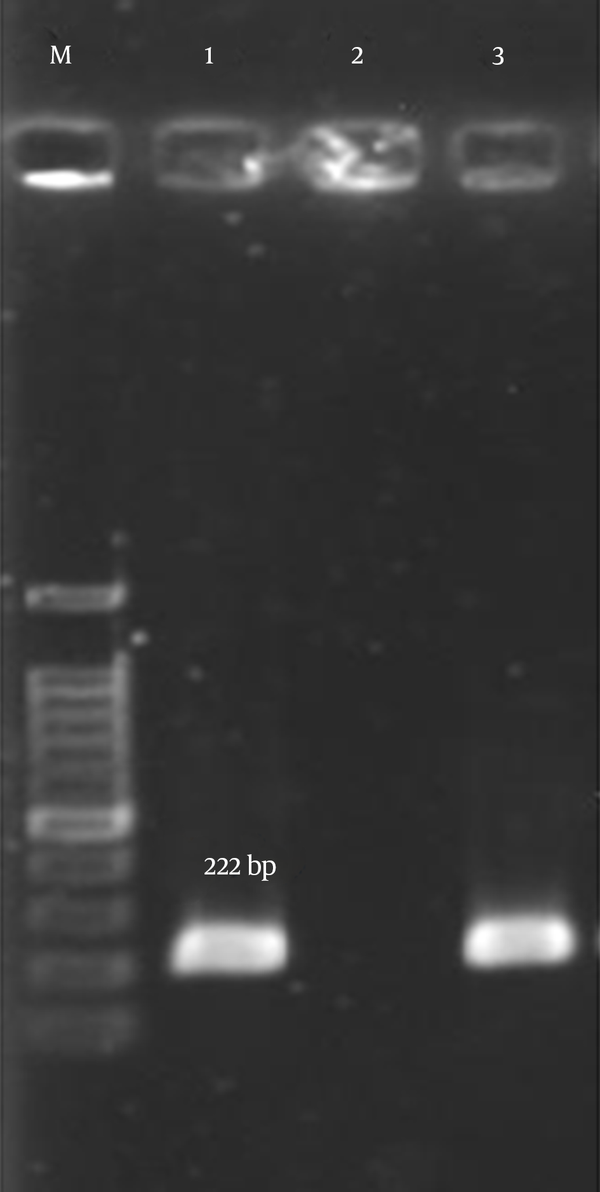
Detection of Genes Encoding aph(3’)-III in CNS Isolates by PCR
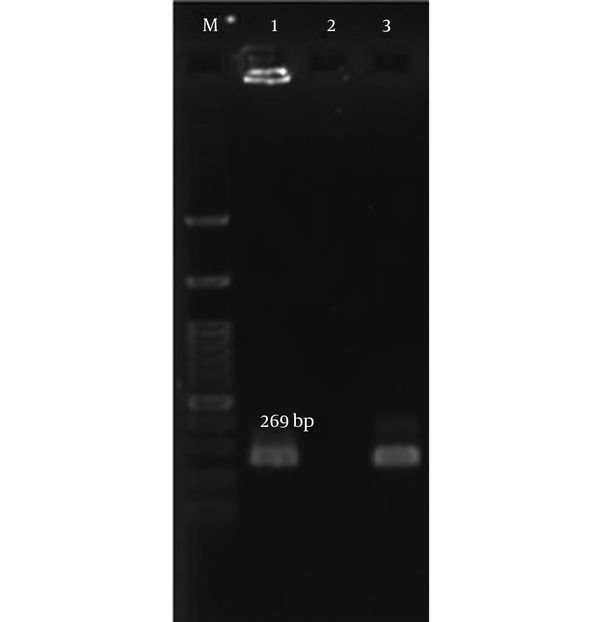
5. Discussion
Antibiotic resistance remains a major threat to public health worldwide (10). Although other studies showed that the most prevalent spices was S. epidermidis (11, 12), the results of the present study showed that S. hominis and S. haemolyticus, as the most important causes of nosocomial infections, had the highest prevalence (10). Insufficient hand hygiene and inadequate disinfection and/or sterilization of medical instruments and surfaces may also be assumed as causes of the distribution of coagulase-negative Staphylococci (CoNS) such as S. hominis and S. haemolyticus in the hospital settings (13). As previously mentioned, excessive consumption of different antibiotics has led to the emergence of multi-drug resistance in the developing countries (14). Aminoglycosides are an important group of antibiotics in the treatment of serious bacterial infections, especially Gram negative bacteria, but the current reports indicated the emergence of resistance to aminoglycosides in CNS isolates in different parts of the world (15).
The methicillin resistance rate of 86% (89/103) among the study CNS isolates was higher than that of reported in Colombia (73%) (16), Egypt (75%) (17), and Brazil (77%) (18), which could be due to the overuse of antibiotics that eradicate MSCNS and facilitate MRCNS colonization. Furthermore, activity in screening, outbreak investigations, and contact tracing regarding methicillin-resistant coagulase-negative Staphylococci (MRCoNS) may differ between Iran and the abovementioned countries. Despite many previous studies, which had shown the more common rate of aac (6’)/aph (2’’) gene (19-21), in the current study, the highest prevalence of aminoglycoside resistance genes was related to ant (4’)-I (87.6%). This finding was similar to the ones reported from Kuwait (88%) (21) and Japan (84.5%) (22). The 2nd most prevalent AMEs gene was aac (6’)-Ie-aph (2’’), which can inactivate gentamicin, kanamycin, tobramycin, neomycin, and amikacin (23). The 3rd gene was aph (3)-IIIa, which inactivates kanamycin and amikacin (23). Although it was found to be the lowest in the current study (66.3%), its amount was higher than reported by other studies such as those of Ghotaslou (19.3%) (15) and Schmitz (14%) (24). Differences between reports from different countries could be due to differences among the isolates and geographical regions. The probable coexistence of all 3 enzymes was detected in the current study isolates, similar to the other 2 researches. They showed that most of the isolates carried the genes for aac (6’)-aph (2’’), ant (4’), and aph (3’), and only a few of them carried genes for single enzymes due to the presence of the same plasmid or transposon encoding the AMEs in CNS species (21). According to the existence of 2 mecA and AMEs genes, many studies showed a correlation between aminoglycoside and methicillin resistance (25, 26).
In general, identification of the species and their resistance patterns contribute to appropriate prescription of antibiotics and accordingly, decrease of antibiotic resistance rate and prevention of widespread resistant genes among CNSs. Because the resistance of CNS to multiple antimicrobial agents varies from one species to another, determining susceptibility to antibiotics for each species allows the researchers to intercept outbreaks of hospital infections caused by CNS. Meanwhile, prescription of antibiotics, including aminoglycoside, should be done with more surveillance. Thus, it seems likely that phenotypic measures, rapid and molecular methods, as well as detection of the susceptibility of bacterial isolates to antimicrobial agents contribute to appropriate treatment of the patients.
Acknowledgements
References
-
1.
Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870-926. [PubMed ID: 25278577]. https://doi.org/10.1128/CMR.00109-13.
-
2.
Oliveira CF, Cavanagh JP, Fredheim EG, Reiter KC, Rieger A, Klingenberg C, et al. Coagulase-negative staphylococci in Southern Brazil: looking toward its high diversity. Rev Soc Bras Med Trop. 2016;49(3):292-9. [PubMed ID: 27384825]. https://doi.org/10.1590/0037-8682-0015-2016.
-
3.
Morgenstern M, Erichsen C, Hackl S, Mily J, Militz M, Friederichs J, et al. Antibiotic Resistance of Commensal Staphylococcus aureus and Coagulase-Negative Staphylococci in an International Cohort of Surgeons: A Prospective Point-Prevalence Study. PLoS One. 2016;11(2). e0148437. [PubMed ID: 26840492]. https://doi.org/10.1371/journal.pone.0148437.
-
4.
Perumal N, Murugesan S, Krishnan P. Distribution of genes encoding aminoglycoside-modifying enzymes among clinical isolates of methicillin-resistant staphylococci. Indian J Med Microbiol. 2016;34(3):350-2. [PubMed ID: 27514959]. https://doi.org/10.4103/0255-0857.188339.
-
5.
Namvar Ebrahimzadeh A, Havaei SA, Azimi L, Lari AR, Rajabnia R. Molecular Characterization of Staphylococcus epidermidis Isolates Collected From an Intensive Care Unit. Arch Pediatr Infect Dis. 2016;5(2).
-
6.
Rahimi F. Characterization of Resistance to Aminoglycosides in Methicillin-Resistant Staphylococcus aureus Strains Isolated From a Tertiary Care Hospital in Tehran, Iran. Jundishapur J Microbiol. 2016;9(1). e29237. [PubMed ID: 27099687]. https://doi.org/10.5812/jjm.29237.
-
7.
Sabzehali F, Gudarzi M, Goudarzi H. Determination of antibiotic resistance in clinical isolates of coagulase negative Staphylococci from hospitalized patients in selected hospitals of Tehran. J Paramed Sci. 2015;6(2).
-
8.
Aghazadeh M, Rezaee MA, Nahaei MR, Mahdian R, Pajand O, Saffari F, et al. Dissemination of aminoglycoside-modifying enzymes and 16S rRNA methylases among acinetobacter baumannii and Pseudomonas aeruginosa isolates. Microb Drug Resist. 2013;19(4):282-8. [PubMed ID: 23577624]. https://doi.org/10.1089/mdr.2012.0223.
-
9.
Cockerill FR. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. Clinical and Laboratory Standards Institute (CLSI); 2011.
-
10.
Krzyminska S, Szczuka E, Dudzinska K, Kaznowski A. Virulence and the presence of aminoglycoside resistance genes of Staphylococcus haemolyticus strains isolated from clinical specimens. Antonie Van Leeuwenhoek. 2015;107(4):857-68. [PubMed ID: 25586730]. https://doi.org/10.1007/s10482-015-0378-6.
-
11.
Farina N, Carpinelli L, Samudio M, Guillen R, Laspina F, Sanabria R, et al. [Clinically significant coagulase-negative staphylococci: most frequent species and virulence factors]. Rev Chilena Infectol. 2013;30(5):480-8. [PubMed ID: 24248161]. https://doi.org/10.4067/S0716-10182013000500003.
-
12.
Shin JH, Kim SH, Jeong HS, Oh SH, Kim HR, Lee JN, et al. Identification of coagulase-negative staphylococci isolated from continuous ambulatory peritoneal dialysis fluid using 16S ribosomal RNA, tuf, and SodA gene sequencing. Perit Dial Int. 2011;31(3):340-6. [PubMed ID: 21454395]. https://doi.org/10.3747/pdi.2010.00073.
-
13.
Hazarika J, Biswas S. Study of Coagulase-Negative Staphylococci Isolated from Clinical Specimens in A Tertiary Care Hospital of North East India. PARIPEX-Indian J Res. 2016;5(7).
-
14.
Anvarinejad M, Sh F, Emamghoreishi F, Hoseini M. Investigating the frequency of multi-drug resistant strains of Escherichia coli isolated from urinary tract infection in children. J Jahrom Univ Med Sci. 2012;9(4):18.
-
15.
Ghotaslou R, Aghazadeh M, Ahangarzadeh Rezaee M, Moshafi MH, Forootanfar H, Hojabri Z, et al. The prevalence of aminoglycoside-modifying enzymes among coagulase negative staphylococci in Iranian pediatric patients. J Infect Chemother. 2014;20(9):569-73. [PubMed ID: 25023717]. https://doi.org/10.1016/j.jiac.2014.05.004.
-
16.
Arias CA. Multicentre surveillance of antimicrobial resistance in enterococci and staphylococci from Colombian hospitals, 2001-2002. J Antimicrob Chemother. 2002;51(1):59-68. https://doi.org/10.1093/jac/dkg002.
-
17.
El Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL. Antimicrobial resistance in Cairo, Egypt 1999-2000: a survey of five hospitals. J Antimicrob Chemother. 2003;51(3):625-30. [PubMed ID: 12615864].
-
18.
Rosa J, Moura J, Palos MAP, Gir E, Reis C, Kipnis A, et al. Deteccao do gene mecA em estafilococos coagulase negativa resistentes a oxacilina isolados da saliva de profissionais da enfermagem. Revista da Sociedade Brasileira de Medicina Trop. 2009;42(4):398-403.
-
19.
Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol Res. 2006;161(1):49-54. [PubMed ID: 16338590]. https://doi.org/10.1016/j.micres.2005.05.002.
-
20.
Emaneini M, Taherikalani M, Eslampour MA, Sedaghat H, Aligholi M, Jabalameli F, et al. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of staphylococci in Tehran, Iran. Microb Drug Resist. 2009;15(2):129-32. [PubMed ID: 19432516]. https://doi.org/10.1089/mdr.2009.0869.
-
21.
Udo EE, Dashti AA. Detection of genes encoding aminoglycoside-modifying enzymes in staphylococci by polymerase chain reaction and dot blot hybridization. Int J Antimicrob Agents. 2000;13(4):273-9. [PubMed ID: 10755241].
-
22.
Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbiol. 2001;39(9):3115-21.
-
23.
Ounissi H, Derlot E, Carlier C, Courvalin P. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive cocci. Antimicrob Agents Chemother. 1990;34(11):2164-8. [PubMed ID: 1963528].
-
24.
Schmitz FJ. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J Antimicrob Chemother. 1999;43(2):253-9. https://doi.org/10.1093/jac/43.2.253.
-
25.
Sorour AE, El-Awady BA, Mukhtar AM. Co-existance of Aminoglycoside Modifying Enzymes (AMEs) genes and mec-A gene among nosocomial isolates of Staphylococcus aureus in Surgical Intensive Care Units in Kasr Al-Ainy hospitals, Cairo University. Int Arab J Antimicrob Agents. 2014;3(4).
-
26.
Vitali LA, Petrelli D, Lamikanra A, Prenna M, Akinkunmi EO. Diversity of antibiotic resistance genes and staphylococcal cassette chromosome mec elements in faecal isolates of coagulase-negative staphylococci from Nigeria. BMC Microbiol. 2014;14:106. [PubMed ID: 24766644]. https://doi.org/10.1186/1471-2180-14-106.