Abstract
Background:
Resistance to rifampin in multidrug-resistant M. tuberculosis (MDR-TB) is one of the major threats to the global public health system.Objectives:
The aim of the present study was to explore the molecular characterization of resistance against rifampin amongst multidrug-resistant (MDR) and extensively drug-resistant (XDR) isolates obtained from patients.Methods:
In this study, we used XDR (n = 6), MDR (n = 9) and rifampin sensitive (n = 39) isolates whose drug susceptibility patterns were identified by proportion method. A simple single-step multiplex-allele specific-polymerase chain reaction (MAS-PCR) assay was optimized to detect the three most frequent mutations in rifampin resistance-determining region (RRDR) fragment of rpoB gene. Additionally, single-strand conformational polymorphism (SSCP)-PCR and Sequencing were utilized.Results:
Out of nine MDR isolates, five were detected by MAS-PCR as rifampin resistant (sensitivity; 55.5%). In comparison with the proportion method, the sensitivity of SSCP for XDR, MDR and rifampin sensitive isolates were 83.3%, 77.7% and 97.4%, respectively. The DNA sequencing revealed that three of the 6 XDR isolates had several silent mutations, one isolate had one silent mutation, one strain had no mutations and only one isolate had mutations at codon 531.Conclusions:
In sum, although the molecular methods are rapid, they are not able to identify resistance against rifampin efficiently.Keywords
Mycobacterium tuberculosis Multidrug-Resistant Extensively Drug-Resistant Rifampin
1. Background
Tuberculosis (TB) remains a deadly infectious disease affecting millions of people worldwide especially in developing countries (1). In Iran, tuberculosis represents a serious public health problem. According to the Iranian ministry of health, the estimated incidence rate of TB was 13 cases per 100,000 persons in 2014 (2). The TB problem has become more severe because of the increase in multidrug-resistant (MDR) M. tuberculosis strains (3).
In recent years, a dramatic increase has been observed in the isolation of extensively drug-resistant (XDR) in M. tuberculosis isolates (4-6). According to the World Health organization (WHO), in 2014 there were an estimated 480,000 new cases of MDR-TB worldwide and approximately 190,000 deaths from MDR-TB (7). With attention to the WHO annual report, the prevalence of MDR-TB in Iran in 2005, 2010 and 2014 has been 0.3%, 0.57% and 0.8%, respectively (7, 8). The report of Masjedi et al. (9) showed that out of 113 MDR isolates from Iranian patients, 12 were XDR. Since emergence of XDR has been reported as a major health threat, which mostly occur among MDR M. tuberculosis, the rapid detection and treatment of drug-resistant TB is a crucial way to prevent XDR-TB (10).
Rifampin is an effective bactericidal agent against M. tuberculosis. Rifampin mediates its bactericidal activity against M. tuberculosis by interacting with DNA dependent RNA polymerase to inhibit transcription and elongation of RNA (11). Since rifampin mono-resistance is extremely rare and the development of isoniazid resistance usually precedes that of rifampin, resistance against rifampin is usually considered to be the MDR marker (12). Therefore, we can prospect the spread of MDR and XDR tuberculosis through detecting the rifampin resistance.
Various methods have been introduced to determine the drug resistance in M. tuberculosis isolates and each method has its own advantages and limitations. An appropriate method should be acceptable regarding its cost, time consumption, accessibility, feasibility, specificity and sensitivity. The proportion method, as a gold standard method, which is based on classical culture, requires 6 to 8 weeks and high level of safety (12, 13).
Another phenotypic method is the Colourimetric technique, this uses reagents such as alamar blue, malachite green and 2,3,5-triphenyltetrazolium chloride (TTC) for detecting drug resistance in M. tuberculosis, which are generally based on the reduction of an oxidation–reduction indicator added to a liquid culture medium after M. tuberculosis has been exposed in vitro to different antibiotics. Resistance is detected by a change in color of the oxidation-reduction indicator (14, 15).
The molecular investigations of rifampin resistance amongst M. tuberculosis have shown that 96% of rifampin-resistant M. tuberculosis strains possess genetic alterations within an 81-bp rifampin resistance-determining region (RRDR) in the rpoB gene, corresponding to codons 507 to 533 (Escherichia coli numbering system) (16). Molecular detection of these changes in the rpoB gene constitutes a reliable method for the rapid detection of rifampin resistance and MDR-TB. Several molecular techniques have been successfully used for the rapid detection of mutations within the rpoB gene, including rpoB sequencing, Multiplex-Allele Specific PCR (MAS-PCR) (12, 13, 17), single strand conformation polymorphism (SSCP)-PCR (18-20), Real-Time PCR (21), commercialized Line Probe Assay (22) and Gene Xpert (23).
MAS-PCR permits simultaneous detection of the most common INH and RIF resistance-associated genetic mutations (13). SSCP-PCR analysis involves amplification by PCR of a segment of the gene encoding for the specific drug target, in which mutations result in an altered pattern (24). The Gene Xpert MTB/RIF assay was an automated molecular test for detection of M. tuberculosis (MTB) and resistance to rifampin. This uses heminested real-time PCR assay to amplify an MTB specific sequence of the rpoB gene (25). Although the molecular methods are rapid, they sometimes show different results compared to phenotypic methods. This difference causes lower specificity and sensitivity for these methods when used for M. tuberculosis susceptibility testing (12, 13).
2. Objectives
The purpose of this study was to evaluate the sensitivity of multiplex allele-specific-PCR, single-stranded conformational polymorphism-PCR and DNA sequencing for detection of rifampin-resistant amongst multidrug resistant and extensively drug-resistant M. tuberculosis isolated from patients in Iran.
3. Methods
3.1. M. tuberculosis Isolates and Drug Susceptibility Testing
In this cross-sectional study, the M. tuberculosis isolates were recovered from patients with active clinical infections (2014 - 2015), all isolates were cultured on the Lowenstein-Jensen medium (Merck Co., Germany) and were identified based on conventional methods including staining by Ziehl Neelsen method, colony morphology, niacin production, nitrate reductase, resistance to thiophen-2-carboxylic acid hydrazide and catalase tests (26). MDR isolates were defined as resistant to at least rifampicin and isoniazid and XDR-TB isolates were defined as MDR-TB plus resistant to at least one fluoroquinolone and a second-line injectable (7). In this study, 9 isolates of M. tuberculosis were MDR, 6 isolates were XDR isolates and 39 isolates were susceptible to rifampin. The reference strain of M. tuberculosis H37RV (ATCC 27294), which is sensitive to rifampin (wild-type strain), was used as control.
Drug susceptibility testing of these isolates against isoniazid, rifampin, streptomycin and ethambutol (Sigma Co., USA) was performed at concentrations of 0.2, 40, 4.0 and 2.0 µg/mL, respectively using the proportion method (as a gold standard method) on Lowenstein–Jensen media (9, 27). Resistance to any of the tested drugs was defined as ≥ 1% growth on drug-containing mediums compared with the control medium. Drug-susceptibility testing against second-line drugs (kanamycin, amikacin, capreomycin, ciprofloxacin, cycloserine, ethionamide and para-aminosalicylic acid [Sigma Co., USA]) was performed using 2 critical proportions of 1% and 10%. All XDR isolates were resistant to all 8 second-line drugs tested.
Genomic DNA from all cultivated isolates and reference strain were extracted by the CTAB-NaCl (Merck Co., Germany) (28).
3.2. Multiplex Allele Specific (MAS)-PCR
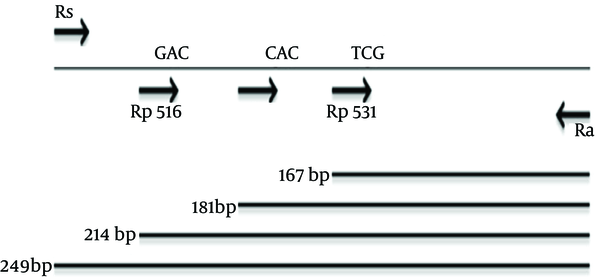
The first pair of primers (forward Rs 5'-GTCGCCGCGATCAAGGA-3' and reverse Ra 5'-TGACCCGCGCGTACAC-3') was used to amplify a 249 bp fragment (12). This fragment corresponding to a portion of rpoB gene (include RRDR fragment) was used for SSCP, sequencing and for PCR control in MAS-PCR. In addition, three allele specific primers targeting codons 516 (5'-GCT GAG CCA ATT CAT GGA-3'), 526 (5'-GTC GGG GTT GAC CCA-3') and 531 (5'-ACA AGC GCC GAC TGT C-3') were used as forward primers (12). Rp516, Rp526 and Rp531 in the rpoB gene were designed for 3' end of mentioned codons as wild type forms of codons (Figure 1). In each MAS-PCR, one allele specific primer (Rp516, Rp526 and Rp531) and the reverse primer (Ra) were used for mutation detection, which generated a short PCR product (214 bp, 181 bp and 167 bp, respectively) from the wild-type gene but failed to amplify from a mutant allele. The control forward primer (Rs) amplified a 249 bp of rpoB gene, which was used in conjugation with the Ra primer as an internal control.
For each MAS-PCR, 3 µL of purified genomic DNA were added to the PCR mixture containing 4 pmol Rs control forward primer, 100 pmol Rp516 or 150 pmol Rp526 or 50 pmol Rp531, 20 pmol reverse primer Ra, 1.5 mM MgCl2, 200 µM deoxynucleoside triphosphates and 2 U of Taq DNA polymeras (final volume of 20 µL) (Fermentase Co., USA). The reaction was performed in a thermo cycler (Eppendorf Co., Germany) under the following program: initial denaturation at 95°C for 10 min, 32 cycles of 95°C for 50 s, 60°C for 40 seconds, 72°C for 40 seconds and final polymerization at 72°C for 10 minutes.
Each isolate was employed in three single tubes with three different primer sets; then the amplified fragments were analyzed in 1.5% agarose gels (Merck Co., Germany) and visualized under UV light.
Primers and PCR Products Length
3.3. Single-Strand Conformational Polymorphism Analysis (SSCP)
Purified genomic DNA templates were amplified for SSCP analysis with Rs and Ra primers in a 20 µL reaction volume containing 1 × PCR mixture (with 1.5 mM MgCl2), 200 mM don’ts, 20 pmol of each primer and 1 U of Taq polymerase (Fermentase Co., USA). Thermocycling condition was performed the same as the MAS-PCR described above.
A 5 µL of amplified DNA was mixed with 10 µL of denaturing buffer (95% formamide, 0.05% xylene cyanole FF and 0.05% bromophenol blue [Merck Co., Germany]), denatured at 95°C for 10 minutes, then chilled on ice for 5 minutes and finally loaded onto 10% acrylamidebisacrylamide (37.5: 1) gels (Merck Co., Germany). Electrophoresis was performed at 120 V for 15 - 18hours in 1 × TBE buffer (Merck Co., Germany) at room temperature. After the electrophoresis, the gel was then stained by a silver staining method (29). H37Rv was used to provide reference wild type patterns on each gel.
3.4. DNA Sequencing
For sequencing, the 249 bp PCR products were directly sequenced by the chain termination method on ABI 377 sequencing machine (GATC Co., Denmark) and the data was analyzed using the MEGA 4 software against M. tuberculosisrpoB reference sequence (GenBank Accession No. NC_000962.2).
3.5. Data Analysis
The sensitivity tests were done using the OpenEpi program. Data was analyzed by the SPSS statistical software version 20.
4. Results
4.1. Multiplex Allele Specific-PCR
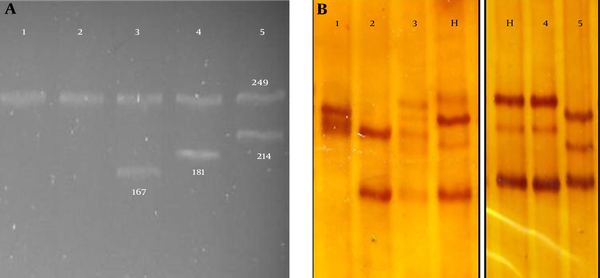
MAS-PCR assay performed on resistant and susceptible clinical isolates indicated that only one of the 39 susceptible isolates showed mutations at codon 531. Amongst nine MDR isolates by proportion method, five were detected by MAS-PCR as rifampin resistant (sensitivity; 55.5%). Amongst these 5 isolates, four isolates had mutations at codon 526 and one isolate had mutations at codon 531. Only 1 out of 6 XDR isolates was found to have mutations at codon 531 in the rpoB gene with a sensitivity of 16.7% (Table 1). MAS-PCR products of some strains with mutations in the 516, 526 and 531 codons of the rpoB gene or without mutations are shown in Figure 2a.
Sensitivity of MAS-PCR, SSCP-PCR and Sequencing
| Susceptibility Pattern (n) | No. of isolates With Detected Mutation by MAS-PCR | Sensitivity of MAS-PCR (%) | No. of Isolates With Detected Mutation by SSCP | Sensitivity of SSCP (%) | No. of Isolates With Detected Mutation by Sequencing | Sensitivity of Sequencing (%) |
|---|---|---|---|---|---|---|
| XDR (6) | 1 | 16.7 | 5 | 83.3 | 5 | 83.3 |
| MDR (9) | 5 | 55.5 | 7 | 77.7 | 8 | 88.9 |
| Rifs (39)a | 1 | 97.4 | 1 | 97.4 | - | - |
4.2. Single Strand Conformation Polymorphism-PCR
The 249 bp fragment of rpoB gene from these isolates and H37Rv strain were amplified successfully in PCR reactions and this fragment containing RRDR was analyzed for mutation by SSCP in comparison with H37Rv strain (Figure 2b). Twelve (5 XDR isolates along with 7 MDR ones) out of 15 rifampin resistant isolates, and one out of 39 rifampin sensitive isolates had different patterns compared to the H37Rv strain. In comparison with the standard proportion method, the sensitivity of SSCP for XDR, MDR and rifampin sensitive isolates were 83.3%, 77.7% and 97.4%, respectively.
(a) MAS PCR Results From Mutant Isolates (1, 2) and Susceptible Isolates for 531 Codon (Line 3), 526 Codon (line 4) and 516 Codon (Line 5) (b) Some PCR-SSCP Electrophoresis Gel Fragment Patterns
4.3. DNA Sequencing
Table 2 represents the mutations, which were detected by sequencing in rifampin resistant isolates. Sequencing of the 249 bp region of the rpoB gene revealed the origin of difference between the results of MAS-PCR and the proportion standard method. There were several silent mutations in three XDR isolates. Moreover, one XDR isolate showed one silent mutation, one isolate without any mutation and one isolate with mutation at codon 531. Eight isolates of MDR showed mutation by sequencing mostly at codon 526.
Mutations Detected in Sequencing in MDR and XDR Isolated from Iranian Patients
| Strain No. | Susceptibility Pattern | No. of Mutation | Mutations Detected in Sequencing | SSCP | MAS-PCR |
|---|---|---|---|---|---|
| R1 | XDR | 7 | GGG(Gly) 523 GGT(Gly); CGA(Arg) 529 CGC(Arg); CCC(Pro) 535 CCG(Pro); CGT(Arg) 542 CGG(Arg); GGG(Gly) 544 GGC(Gly); TGC(Cys) 559 TGT(Cys); GAA(Glu) 562 GAG(Glu) | + | W T |
| R13 | XDR | 15 | CAA(Gln) 513 CAG(Gln) TCG(Ser) 522 TCC(Ser) GGG(Gly) 523 GGT(Gly); TTG(Leu) 524 CTG(Leu); CGA(Arg) 529 CGC(Arg); CTG(Leu) 530 CTC(Leu); GGG(Gly) 534 GGC(Gly); TCA(Ser) 539 TCC(Ser); CGT(Arg) 542 CGC(Arg); GGG(Gly) 544 GGC(Gly); CGC(Arg) 548 CGT(Arg); CTG(Val) 550 GTC(Val); CCG(Pro) 552 CCC(Pro); CCT(Pro) 564 CCG(Pro); GGG(Gly) 566 GGT(Gly) | + | WT |
| R21 | MDR | 1 | GAA(Glu) 562 GAG(Glu) | + | WT |
| R23 | MDR | 1 | CAC(His) 526 AAC(Asn) | + | 526 His |
| R29 | MDR | 1 | CAC(His) 526 AAC(Asn) | - | 526 His |
| R45 | XDR | 12 | TCG(Ser) 522 TCC(Ser); GGG(Gly) 523 GGT(Gly); TTG(Leu) 524 CTG(Leu); CGA(Arg) 529 CGC(Arg) CTG(Leu) 530 CTC(Leu); GGG(Gly) 534 GGC(Gly); TCA(Ser) 539 TCC(Ser); CGT(Arg) 542 CGC(Arg); GGG(Gly) 544 GGC(Gly); CGC(Arg) 548 CGT(Arg); CTG(Val) 550 GTC(Val); GAA(Glu) 562 GAG(Glu) | + | WT |
| R48 | MDR | 1 | CAC(His) 526 AAC(Asn) | + | 526 His |
| R52 | MDR | 1 | TCG(Ser) 531 TTG(Leu) | + | 531 Ser |
| R53 | MDR | 1 | CTG(Leu) 533 CCG(Pro) | + | WT |
| R66 | XDR | - | No Mutation | - | WT |
| R67 | XDR | 1 | TCG(Ser) 531CAG(Gln) | + | 531 Ser |
| R69 | MDR | 1 | CAC(His) 526TAC(Tyr) | + | 526 His |
| R71 | MDR | - | No Mutation | - | WT |
| R72 | MDR | 2 | GAC(Asp) 516 TAC(Tyr); CAC(His) 526 AAC(Asn) | + | WT; 526 His |
| R74 | XDR | 1 | GGG(Gly) 544 GGC(Gly) | + | WT |
5. Discussion
Resistance to first line antibiotics in multidrug-resistant M. tuberculosis is one of the major threats to global public health systems (30). In recent years, extensively drug-resistant tuberculosis has emerged worldwide as having even more sever threats (6, 31). The WHO has reported that XDR-TB has been found in 105 countries (7). In Asia, some countries have reported the incidence of XDR-TB. In Iran and Hong Kong, 10.6% and 9.2% of MDR isolates were, respectively, XDR ones (9, 32). In china, thirteen isolates amongst 1,926 clinical isolates were XDR (33).
According to the recommendations of the WHO Global Task Force on XDR-TB, one of the important ways to stop the incidence of drug resistant TB is to strengthen laboratory diagnostic services to ensure rapid and accurate drug-susceptibility testing (34). Therefore, rapid strengthen detection of mycobacteria with prompt anti-mycobacterial susceptibility testing is essential to control tuberculosis and prevent the spread of resistant pathogens.
In current works, amongst 6 XDR isolates, only 1 isolate was found to have mutations at codon 531 in the rpoB gene by MAS-PCR, which was confusing. The DNA sequencing revealed that three of the 6 XDR isolates had several silent mutations, one isolate had one silent mutation, one isolate did not possess any mutation and only 1 had mutations at codon 531. Such findings concerning XDR isolates with several silent mutations have not been reported up to now. The study of Sun and Chao among 13 XDR isolates from China revealed that 9 isolates had mutation in codon 531 (S531L) (33). In another study, Bahremand et al. (35) reported multiple mutations, which correlated with high level rifampin resistance in MDR isolates. While previous studies have reported more than 90% sensitivity for rifampin susceptibility testing using MAS-PCR (12, 16). In our study, the sensitivity of the MAS-PCR for rifampin resistant detection among MDR isolates were 55.5%, a finding which must be considered with caution as we performed our study on a small number of isolates. In contrast, mutations tracking in RRDR region of XDR isolates revealed such a noticeable finding that they can propose some relation between silent mutations and extensively drug-resistant ones. A major limitation of the molecular genetic detection of drug resistance is that these tests only detect known mutations. Since not all mutations conferring resistance to anti-TB drugs are known and the prevalence of mutations may vary by geographic area, identification of resistance-associated mutations can only be informative. However, lack of mutation in the target sequence must be interpreted with caution. Our findings emphasize the importance of this caution especially for XDR-isolates.
In the present study, the SSCP-PCR of RRDR revealed that out of 15 rifampin resistant isolates, 12 had different patterns compared to strain H37Rv. Similar results have also been reported by others (36, 37). Sheikholslami and colleagues (24) reported that sensitivity of PCR-SSCP method was 70.8%. This result is similar to our findings (sensitivity, 77.7%), but in the other study performed by Imani et al. (38) the sensitivity of PCR-SSCP for MDR isolate was 33%.
In the study of Sahebi et al. (26) the sensitivity of real-time PCR in detecting rifampin resistance in comparison to the standard proportion test, was 83.3%. Similar to this result, in this work, the sensitivity of SSCP-PCR and DNA sequencing in detecting rifampin resistance in XDR isolates was 83.3% and 83.3%, respectively.
The diagnostic value of SSCP-PCR in our study, similar to the one reported in other reports, was greater than that of MAS-PCR. This greater value can be explained as SSCP-PCR examines the whole product, while MAS-PCR only looks for predefined point mutations. Although SSCP-PCR has higher sensitivity than MAS-PCR, it is not a complete and acceptable method for detection of drug resistance.
The GeneXpert MTB/RIF assay is a novel integrated diagnostic device that performs sample processing and heminested real-time PCR analysis in a single hands-free step for the diagnosis of tuberculosis and rapid detection (within 2 hours) of rifampin resistance in clinical specimens (23). According to the Steingart and coworkers (39) report, the Gene Xpert method sensitivity estimates ranged from 58% to 100% and its specificity estimates ranged from 86% to 100%.
Currently, the proportion method is using as a gold standard technique for detection of rifampin resistant M. tuberculosis in the reference laboratory. In our study, sensitivity and specificity of proportion method was 100% and 100%, respectively.
In conclusion, even though the molecular methods are rapid, they are not able to identify resistance against rifampin efficiently. In other words, the molecular tests cannot completely replace the culture based phenotypic susceptibility test because of the limitation mentioned above. Therefore, it is necessary to choose a rapid method, which has an acceptable sensitivity for all M. tuberculosis isolates.
Acknowledgements
References
-
1.
Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368(8):745-55. [PubMed ID: 23425167]. https://doi.org/10.1056/NEJMra1200894.
-
2.
Regional office for the eastern mediterranean: Islamic Republic of Iran profile:World Health Organization. World Health Organization,; 2015. Available from: http://www.who.int/tb/country/data/profiles/en/.
-
3.
Palomino JC, Martin A. Drug Resistance Mechanisms in Mycobacterium tuberculosis. Antibiotics (Basel). 2014;3(3):317-40. [PubMed ID: 27025748]. https://doi.org/10.3390/antibiotics3030317.
-
4.
Jassal M, Bishai WR. Extensively drug-resistant tuberculosis. Lancet Infect Dis. 2009;9(1):19-30. [PubMed ID: 18990610]. https://doi.org/10.1016/S1473-3099(08)70260-3.
-
5.
Madariaga MG, Lalloo UG, Swindells S. Extensively drug-resistant tuberculosis. Am J Med. 2008;121(10):835-44. [PubMed ID: 18823850]. https://doi.org/10.1016/j.amjmed.2008.04.015.
-
6.
Ahmed I, Jabeen K, Inayat R, Hasan R. Susceptibility testing of extensively drug-resistant and pre-extensively drug-resistant Mycobacterium tuberculosis against levofloxacin, linezolid, and amoxicillin-clavulanate. Antimicrob Agents Chemother. 2013;57(6):2522-5. [PubMed ID: 23507286]. https://doi.org/10.1128/AAC.02020-12.
-
7.
Global tuberculosis report. Geneva: World Health Organization; 2015. Available from: http://www.who.int/tb/publications/global_report/en/.
-
8.
Global tuberculosis control: WHO report. WHO; 2011. Available from: http://apps.who.int/iris/bitstream/10665/44728/1/9789241564380_eng.pdf.
-
9.
Masjedi MR, Farnia P, Sorooch S, Pooramiri MV, Mansoori SD, Zarifi AZ, et al. Extensively drug-resistant tuberculosis: 2 years of surveillance in Iran. Clin Infect Dis. 2006;43(7):841-7. [PubMed ID: 16941364]. https://doi.org/10.1086/507542.
-
10.
Jacobson KR, Tierney DB, Jeon CY, Mitnick CD, Murray MB. Treatment outcomes among patients with extensively drug-resistant tuberculosis: systematic review and meta-analysis. Clin Infect Dis. 2010;51(1):6-14. [PubMed ID: 20504231]. https://doi.org/10.1086/653115.
-
11.
Blanchard JS. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem. 1996;65:215-39. [PubMed ID: 8811179]. https://doi.org/10.1146/annurev.bi.65.070196.001243.
-
12.
Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Allele-specific rpoB PCR assays for detection of rifampin-resistant Mycobacterium tuberculosis in sputum smears. Antimicrob Agents Chemother. 2003;47(7):2231-5. [PubMed ID: 12821473].
-
13.
Chia BS, Lanzas F, Rifat D, Herrera A, Kim EY, Sailer C, et al. Use of multiplex allele-specific polymerase chain reaction (MAS-PCR) to detect multidrug-resistant tuberculosis in Panama. PLoS One. 2012;7(7):40456. [PubMed ID: 22792333]. https://doi.org/10.1371/journal.pone.0040456.
-
14.
Palomino JC, Martin A, Portaels F. Rapid drug resistance detection in Mycobacterium tuberculosis: a review of colourimetric methods. Clin Microbiol Infect. 2007;13(8):754-62. [PubMed ID: 17378933]. https://doi.org/10.1111/j.1469-0691.2007.01698.x.
-
15.
Mohammadzadeh A, Farnia P, Ghazvini K, Behdani M, Rashed T, Ghanaat J. Rapid and low-cost colorimetric method using 2,3,5-triphenyltetrazolium chloride for detection of multidrug-resistant Mycobacterium tuberculosis. J Med Microbiol. 2006;55(Pt 12):1657-9. [PubMed ID: 17108268]. https://doi.org/10.1099/jmm.0.46442-0.
-
16.
Fan XY, Hu ZY, Xu FH, Yan ZQ, Guo SQ, Li ZM. Rapid detection of rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis isolates in shanghai by using the amplification refractory mutation system. J Clin Microbiol. 2003;41(3):993-7. [PubMed ID: 12624020].
-
17.
Gupta A, Prakash P, Singh SK, Anupurba S. Rapid genotypic detection of rpoB and katG gene mutations in Mycobacterium tuberculosis clinical isolates from Northern India as determined by MAS-PCR. J Clin Lab Anal. 2013;27(1):31-7. [PubMed ID: 23325741]. https://doi.org/10.1002/jcla.21558.
-
18.
Kim BJ, Kim SY, Park BH, Lyu MA, Park IK, Bai GH, et al. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997;35(2):492-4. [PubMed ID: 9003625].
-
19.
Grutzmacher LK, Dalmarco EM, Blatt SL, Cordova CM. Drug resistance of Mycobacterium tuberculosis strains in southern Brazil. Rev Soc Bras Med Trop. 2012;45(1):95-9. [PubMed ID: 22370836].
-
20.
Miyata M, Pavan FR, Sato DN, Marino LB, Hirata MH, Cardoso RF, et al. Drug resistance in Mycobacterium tuberculosis clinical isolates from Brazil: phenotypic and genotypic methods. Biomed Pharmacother. 2011;65(6):456-9. [PubMed ID: 21880463]. https://doi.org/10.1016/j.biopha.2011.04.021.
-
21.
Kocagoz T, Saribas Z, Alp A. Rapid determination of rifampin resistance in clinical isolates of Mycobacterium tuberculosis by real-time PCR. J Clin Microbiol. 2005;43(12):6015-9. [PubMed ID: 16333091]. https://doi.org/10.1128/JCM.43.12.6015-6019.2005.
-
22.
Soudani A, Hadjfredj S, Zribi M, Masmoudi A, Messaoud T, Tiouri H, et al. Characterization of Tunisian Mycobacterium tuberculosis rifampin-resistant clinical isolates. J Clin Microbiol. 2007;45(9):3095-7. [PubMed ID: 17652484]. https://doi.org/10.1128/JCM.00643-07.
-
23.
Zeka AN, Tasbakan S, Cavusoglu C. Evaluation of the GeneXpert MTB/RIF assay for rapid diagnosis of tuberculosis and detection of rifampin resistance in pulmonary and extrapulmonary specimens. J Clin Microbiol. 2011;49(12):4138-41. [PubMed ID: 21956978]. https://doi.org/10.1128/JCM.05434-11.
-
24.
Sheikholslami MF, Farnia P, Tabarsi P, Aghali Merza M, Valiollah Pour Amiri M, Mohammadi F. Comparison of polymerase chain reaction single-strand conformation polymorphism with DNA sequencing to detect drug resistance of Mycobacterium tuberculosis isolates. Iran J Clin Infect Dis. 2011;6(2):66-70.
-
25.
Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363(11):1005-15. [PubMed ID: 20825313]. https://doi.org/10.1056/NEJMoa0907847.
-
26.
Sahebi L, Khalil Ansarin K, Monfaredan A, Farajnia S, Nili S, Khalili M. Rapid Detection of Rifampicin- and Isoniazid-Resistant Mycobacterium tuberculosis Using Real-Time PCR. Jundishapur J Microbiol.
-
27.
Anti-tuberculosis drug resistance in the world: third global report. Geneva: World Health Organization; 2004. Available from: http://apps.who.int/iris/bitstream/10665/43103/1/9241562854.pdf.
-
28.
van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29(11):2578-86. [PubMed ID: 1685494].
-
29.
Temesgen Z, Satoh K, Uhl JR, Kline BC, Cockerill F3. Use of polymerase chain reaction single-strand conformation polymorphism (PCR-SSCP) analysis to detect a point mutation in the catalase-peroxidase gene (katG) of Mycobacterium tuberculosis. Mol Cell Probes. 1997;11(1):59-63. [PubMed ID: 9076716]. https://doi.org/10.1006/mcpr.1996.0077.
-
30.
Khoshneviszadeh M, Edraki N, Javidnia K, Alborzi A, Pourabbas B, Mardaneh J, et al. Synthesis and biological evaluation of some new 1,4-dihydropyridines containing different ester substitute and diethyl carbamoyl group as anti-tubercular agents. Bioorg Med Chem. 2009;17(4):1579-86. [PubMed ID: 19162489]. https://doi.org/10.1016/j.bmc.2008.12.070.
-
31.
Shah NS, Wright A, Bai GH, Barrera L, Boulahbal F, Martin-Casabona N, et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007;13(3):380-7. [PubMed ID: 17552090]. https://doi.org/10.3201/eid1303.061400.
-
32.
Kam KM, Yip CW. Surveillance of Mycobacterium tuberculosis susceptibility to second-line drugs in Hong Kong, 1995-2002, after the implementation of DOTS-plus. Int J Tuberc Lung Dis. 2004;8(6):760-6. [PubMed ID: 15182147].
-
33.
Sun Z, Chao Y, Zhang X, Zhang J, Li Y, Qiu Y, et al. Characterization of extensively drug-resistant Mycobacterium tuberculosis clinical isolates in China. J Clin Microbiol. 2008;46(12):4075-7. [PubMed ID: 18945837]. https://doi.org/10.1128/JCM.00822-08.
-
34.
WHO Global Task Force outlines measures to combat XDR TB worldwide. World Health Organization; 2006. Available from: http://www.who.int/mediacentre/news/notes/2006/np29/en/index.html.
-
35.
Bahrmand AR, Titov LP, Tasbiti AH, Yari S, Graviss EA. High-level rifampin resistance correlates with multiple mutations in the rpoB gene of pulmonary tuberculosis isolates from the Afghanistan border of Iran. J Clin Microbiol. 2009;47(9):2744-50. [PubMed ID: 19721079]. https://doi.org/10.1128/JCM.r00548-09.
-
36.
Cheng X, Zhang J, Yang L, Xu X, Liu J, Yu W, et al. A new Multi-PCR-SSCP assay for simultaneous detection of isoniazid and rifampin resistance in Mycobacterium tuberculosis. J Microbiol Methods. 2007;70(2):301-5. [PubMed ID: 17543399]. https://doi.org/10.1016/j.mimet.2007.05.002.
-
37.
Isfahani BN, Tavakoli A, Salehi M, Tazhibi M. Detection of rifampin resistance patterns in Mycobacterium tuberculosis strains isolated in Iran by polymerase chain reaction-single-strand conformation polymorphism and direct sequencing methods. Mem Inst Oswaldo Cruz. 2006;101(6):597-602. [PubMed ID: 17072470].
-
38.
Imani F, Babak F, Fazlollah MS, Nematollah JJ. Rapid detection of MDR-Mycobacterium tuberculosis using modified PCR-SSCP from clinical Specimens. Asian Pac J Trop Biomed. 2014;4(Suppl 1):165-70. [PubMed ID: 25183075]. https://doi.org/10.12980/APJTB.4.2014C1186.
-
39.
Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;(1):9593. [PubMed ID: 24448973]. https://doi.org/10.1002/14651858.CD009593.pub3.