Abstract
Background:
Estimated as the second or third most prevalent respiratory pathogen in the pediatric population, routine testing for human metapneumovirus (hMPV) can have a pivotal impact on children’s clinical outcome.Objectives:
This cross-sectional analytical study aimed to determine the efficiency of direct fluorescent antibody (DFA) assay as a rapid tool for the diagnosis of hMPV infection as compared to real time reverse transcriptase polymerase chain reaction (rRT-PCR). In the meantime, we endeavored to analyze the clinical features in hMPV patients.Methods:
A total of 50 children aged ≤ 24 months presenting with manifestations of acute respiratory tract infection (ARTI) at El-Mounira pediatric university hospital, Cairo university were enrolled in the study. Nasopharyngeal aspirates (or endotracheal aspirates in intubated children) were examined with the DFA assay as well as rRT-PCR as a gold standard for the detection and quantification of hMPV.Results and conclusion:
Human MPV was detected in two cases by DFA and in four cases by rRT-PCR among hospitalized children with ARTIs. The DFA assay proved to be a highly specific test, yet with low sensitivity when compared to rRT-PCR. Most of hMPV-infected cases presented during the winter season, with January and February exhibiting the highest hMPV activity. Pneumonia was the most common presentation of ARTIs in hMPV-infected patients. Direct evaluation of respiratory specimens by DFA provides rapid results with low cost and a subsequent early medical management. However, its use should be restricted as a first-line approach, and a confirmatory test would be needed for a definite diagnosis.Keywords
Direct Fluorescent Antibody Assay Polymerase Chain Reaction Human Metapneumovirus Pneumonia Infants
1. Background
Since its first description in 2001, hMPV has been detected in all continents independent of the economic status of different countries (1-3). The prevalence of hMPV in children with acute respiratory tract infection (ARTI) has been estimated as 5% to 15% (4), making it the 2nd or 3rd most prevalent pathogen in children with ARTI, with only human respiratory syncytial virus (hRSV) and possibly rhinovirus being more prevalent (5, 6).
For accurate diagnosis of hMPV infections, four principal methods can be applied: virus isolation via culture, serological tests, RNA detection by reverse transcriptase-polymerase chain reaction (RT-PCR), as well as antigen detection (7, 8). Virus isolation using cell cultures is validated as the “gold standard”, though tedious due to slow viral growth and subtle cytopathic effects without apparent syncytium formation. Serological tests can be of importance for retrospective discrimination between primary infection and reinfection; however, the antibody response in the acute phase is not helpful in the diagnosis of hMPV infections (9). Therefore, RT-PCR is regarded as the most sensitive and specific tool for the detection of hMPV (9, 10). Nonetheless, RT-PCR can be performed only in equipped laboratories and requires > 6 hours to yield results (8). Meanwhile, DFA staining of respiratory epithelial cells present in nasopharyngeal aspirate (NPA) using a virus-specific monoclonal antibody (MAb) is a rapid diagnostic tool for conventional respiratory viruses and is easily performed in clinical virology laboratories (4).
2. Objectives
We endeavored to compare the sensitivity and specificity of the DFA assay versus rRT-PCR in the diagnosis of hMPV infection. In the meantime, we analyzed the clinical features of hMPV infection during the respiratory disease season.
3. Methods
3.1. Ethical Consideration
Before commencement of the study, approval of the protocol was obtained from the ethics committee of the department of microbiology and immunology, Cairo University.
3.2. Population of Study and Disease Condition
This cross-sectional analytical study was conducted on infants presenting with ARTI at El-Mounira pediatric university hospital, Cairo university. A total of 50 infants were selected using non-random purposive sampling. The study was done over a period of six months, from December 2014 through May 2015. Prior to enrollment in the study, an informed written consent from the parents or guardians was obtained after detailed explanation of the study nature. All patients participating in the present study were subjected to history taking and clinical examination for signs of ARTI.
3.3. Inclusion Criteria
Children aged ≤ 24 months presenting with any combination of the following manifestations, cough, difficulty in breathing, fever, chest indrawing, or rapid breathing (≥ 50 breaths/minute in children aged 2 - 11 months or ≥ 40 breaths/minute in children aged 12 - 24 months), were included in the study (11).
3.4. Exclusion Criteria
Patients were excluded if they were < 2 months or > 2 years of age, unable or unwilling to participate according to the parent's or guardian's will or presenting >7 days after the onset of symptoms (8).
3.5. Specimen Collection, Transport and Processing
• From each patient, 1 - 2 mL of nasopharyngeal secretions were aspirated through a catheter fitted to an electric suction device (12). Endotracheal aspirate (ETA) was obtained in intubated patients.
• The catheter was flushed with 3 mL of viral transport medium (MicroTest M4 medium; Remel, Lenexa, KS) (13).
• Upon arrival to the laboratory, samples were mixed via pulse-vortexing and divided into two aliquots; one aliquot was used within 72 hours for the DFA at the virology unit of the medical microbiology and immunology department, Cairo University and the second was kept at -70°C until further testing by rRT-PCR at the molecular biology unit of the medical biochemistry department, Cairo university.
3.6. Detection of Human Metapneumovirus
3.6.1. Direct Fluorescent Antibody assay
This was carried out using light diagnostics human metapneumovirus DFA assay (Millipore, Billerica, MA). The reaction was considered positive when a characteristic granular bright apple-green fluorescence was visualized in the cytoplasm and/or the nucleus. While if a dull red fluorescence was detected, it was considered negative (14).
3.6.2. Real Time Reverse Transcriptase-Polymerase Chain Reaction
Viral RNA was extracted using QIAamp viral RNA mini kit (Qiagen, Germany) as per the manufacturers’ instructions. Both complementary DNA synthesis and PCR were performed in a single tube on the StepOnePlus Real-Time PCR system (applied biosystems, CA, USA). Positive control and negative control (RNAse/DNAse free water) were used in every PCR run (15). The following kits were used according to manufacturer’s instructions.
• Primer design genesig advanced kit (PrimerDesign Ltd, Southampton, Hants, UK) that detects N gene of hMPV.
• PrimerDesign oasig lyophilised OneStep qRT-PCR Mastermix kit (PrimerDesign Ltd, Southampton, Hants, UK).
3.7. Statistical Analysis
All statistical analyses were performed using microsoft excel 2007 (Microsoft Corporation, USA) and SPSS (statistical package for the social science; SPSS Inc., USA) version 16 for microsoft windows. Data were statistically described as range, mean ± standard deviation (SD), median, frequency and percentages. Comparison of age, total leukocytic count, lymphocyte count and absolute neutrophil count was carried out using Mann Whitney U test. In order to compare categorical data, Chi square (χ2) test was performed. Fisher’s exact test was applied when the expected frequency was less than 5. A probability P value of < 0.05 was considered statistically significant. Kappa statistics were used to compute the measure of agreement between DFA and rRT-PCR.
4. Results
From December 2014 through May 2015, fifty pediatric patients were enrolled in this study. Using DFA, two samples (4%) were positive, whereas four samples (8%) tested positive for hMPV by rRT-PCR.
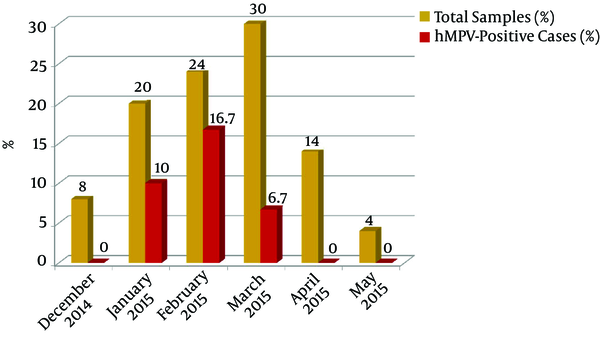
The hMPV infection rate showed no statistical significant difference regarding age or gender (Table 1). Our results revealed that hMPV infections occurred during January through March, with a peak during February (Figure 1).
Age and Gender Distribution Among Human Metapneumovirus (Hmpv) Positive Cases
| Parameter | Total 50, (100%) | hMPV-Positive 4, (8%) | hMPV-Negative 46, (92%) | P Value |
|---|---|---|---|---|
| Age, mo, mean ± SD | 9.34 ± 6.57 | 13.5 ± 8.66 | 8.89 ± 6.36 | > 0.05 |
| Age group, mo | > 0.05 | |||
| < 12 | 35 (70%) | 1 (2.9%) | 34 (97.1%) | |
| 12 - 24 | 15 (30%) | 3 (20%) | 12 (80%) | |
| Gender | > 0.05 | |||
| Male | 27 (54%) | 2 (7.4%) | 25 (92.6%) | |
| Female | 23 (46%) | 2 (8.7%) | 21 (91.3%) |
Seasonal Distribution of Human Metapneumovirus (Hmpv) Positive Cases from December 2014 Through May 2015 at El-Mounira Pediatric University Hospital, Cairo University
Considering the medical history, one hMPV-positive infant had congenital cardiac disease and another one had bronchial asthma (Table 2). A statistically significant difference was observed regarding the lymphocytic count of hMPV-positive cases and hMPV-negative cases. All patients with hMPV infection had respiratory distress with cough as a symptom (Table 3). Antibiotics, corticosteroids, and bronchodilators were the medications mostly offered (100%, 100%, and 75% respectively).
Medical History Characteristics of Human Metapneumovirus (Hmpv)-Infected Patients
| Characteristic | Total 50, (100%) | hMPV-Positive, 4 (8%) | hMPV-Negative, 46 (92%) | P Value |
|---|---|---|---|---|
| Premature birth | 8 (16%) | 1 (25%) | 7 (15.2%) | > 0.05 |
| Breast feeding | 33 (66%) | 3 (75%) | 30 (65.2%) | > 0.05 |
| Similar condition | 20 (40%) | 2 (50%) | 18 (39.1%) | > 0.05 |
| Underlying conditions: | ||||
| Cardiac | 13 (26%) | 1 (25%) | 12 (26.1%) | > 0.05 |
| Bronchial asthma | 11 (22%) | 1 (25%) | 10 (21.7%) | > 0.05 |
Clinical Characteristics of Cases with Human Metapneumovirus (Hmpv) Infection
| Characteristics | Total, N = 50 | hMPV-Positive, N = 4 | hMPV-Negative, N = 46 | P Value |
|---|---|---|---|---|
| Clinical signs and symptoms | ||||
| Respiratory distress | 50 (100%) | 4 (100%) | 46 (100%) | Cannot be computed |
| Cough | 43 (86%) | 4 (100%) | 39 (84.8%) | > 0.05 |
| Wheezing | 34 (68%) | 2 (50%) | 32 (69.6%) | > 0.05 |
| Fever | 32 (64%) | 2 (50%) | 30 (65.2%) | > 0.05 |
| Rhinorrhea | 22 (44%) | 2 (50%) | 20 (43.5%) | > 0.05 |
| Clinical diagnosis | ||||
| Pneumonia | 45 (90%) | 3 (75%) | 42 (91.3%) | > 0.05 |
| Bronchiolitis | 6 (12%) | 1 (25%) | 5 (10.9%) | > 0.05 |
| Asthma | 3 (6%) | 1 (25%) | 2 (4.3%) | > 0.05 |
| Pharyngitis | 7 (14%) | 0 (0%) | 7 (15.2%) | > 0.05 |
| Chext X Ray (CXR) | ||||
| Normal | 2 (4%) | 0 (0%) | 2 (4.3%) | > 0.05 |
| Abnormal | 48 (96%) | 4 (100%) | 44 (95.7%) | > 0.05 |
| Complete Blood Count (CBC) | ||||
| Total leukocytic count, × 109 cells/La | 9.46 ± 3.43 | 9.4 ± 3.22 | 9.46 ± 3.48 | > 0.05 |
| Lymphocytic count(%)a | 55.48 ± 13.19 | 67.75 ± 5.56 | 54.41 ± 13.51 | 0.04b |
| Hospital course | ||||
| Admission to PICU | 10 (20%) | 1 (25%) | 9 (19.6%) | > 0.05 |
| Mechanical ventilation | 8 (16%) | 0 | 8 (17.4%) | > 0.05 |
| Length of stay, da | 10.32 ± 14.26 | 8.5 ± 6.35 | 10.48 ± 14.78 | > 0.05 |
| Complication with acute otitis media (AOM) | 2 (4%) | 1 (25%) | 1 (1.6%) | > 0.05 |
| Outcome | ||||
| Death | 2 (4%) | 0 (0%) | 2 (4.3%) | > 0.05 |
| Discharge | 48 (96%) | 4 (100%) | 44 (95.7%) | > 0.05 |
4.1. Human Metapneumovirus (Hmpv) Sample Description
All hMPV positive cases were recovered from NPA, while ETA was negative by both DFA and rRT-PCR assays (Table 4).
Human Metapneumovirus (Hmpv)-Positive Samples Among the Specimen Types Tested
| Total 50, (100%) | HMPV-positive, 4 (8%) | HMPV-negative, 46 (92%) | P Value | |
|---|---|---|---|---|
| Sample type | ||||
| NPA | 45 (90%) | 4 (8.9%) | 41 (91.1%) | > 0.05 |
| ETA | 5 (10%) | 0 | 5 (100%) | > 0.05 |
| Delay between the onset of symptoms and sample collection, da | 3.24 ± 1.04 | 3 ± 0.8 | 3.26 ± 1.06 | > 0.05 |
4.2. Results of the Direct Fluorescent Antibody assay for Human Metapneumovirus
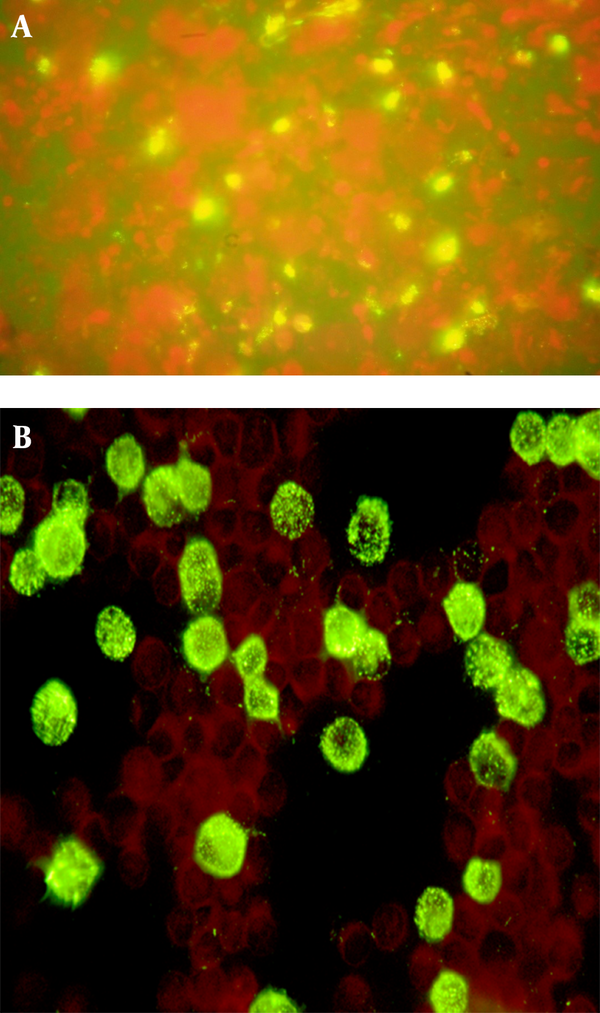
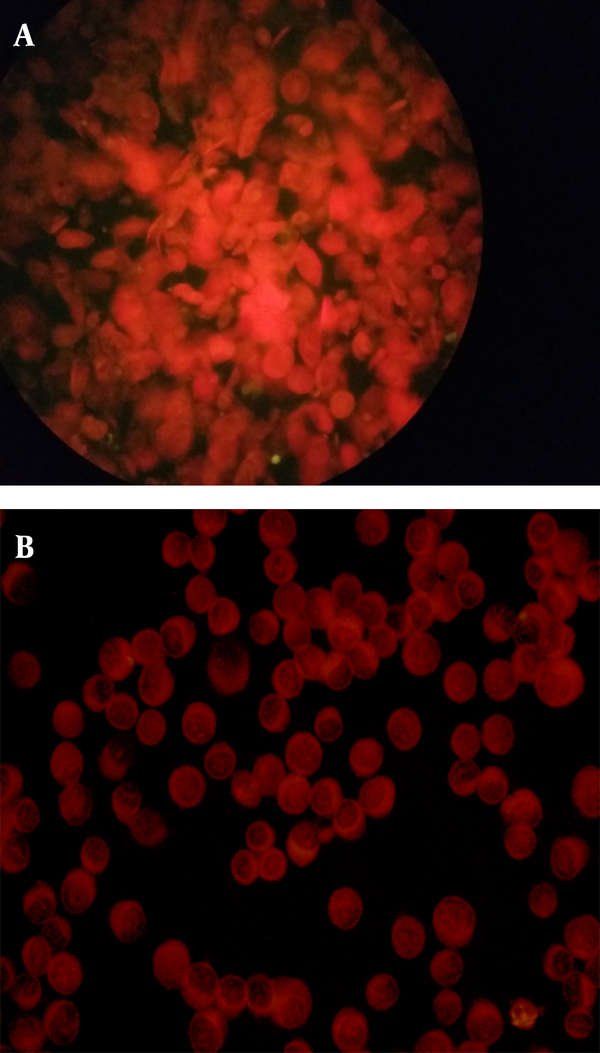
Human metapneumovirus was detected by the DFA assay in two (4%) samples (Figures 2A and 2B), while the remaining 48 samples (96%) were negative (Figures 3A and 3B).
A, positive sample for Human Metapneumovirus (Hmpv) by the direct fluorescent antibody assay showing cells with bright apple green fluorescence (× 400); B, positive control slide containing LLC-MK2 cells infected with hMPV showing the characteristic granular bright apple-green fluorescence (× 400).
A, negative sample for Human Metapneumovirus (Hmpv) by DFA assay showing cells with no fluorescence and stained red due to the Evans Blue counter stain (× 400). B, negative control slide containing non-infected LLC-MK2 cells where no fluorescence was detected and uninfected cells stain dull red (× 400).
4.3. Results of rRT-PCR for Human Metapneumovirus (Hmpv)
Four specimens (8%) were positive for hMPV by rRT-PCR. Out of them, two samples were positive by the DFA assay. Reverse transcription PCR results were expressed as hMPV copies per milliliter of original sample. The numbers of hMPV copies/mL in the four PCR positive samples ranged from 500 to 2 × 104. The sample with the highest template count showed an amplification curve with cycle threshold (CT) value at cycle 25.9 with a copy number of 2 × 104 copies/mL (DFA positive). The second one showed an amplification curve with CT value at cycle 26.9 and the copy number was 1 × 104 copies/mL (DFA negative). The third one showed an amplification curve with CT value at cycle 27.6 and the copy number was 6150 copies/mL (DFA positive). The sample with the least template count showed an amplification curve with CT value at cycle 30.6 and the copy number was 500 copies/mL (DFA negative).
Statistical correlation between the results of rRT-PCR as a standard method with the DFA assay was done as shown in Table 5. Kappa value (statistical measurement of agreement) was < 0.001, denoting a significant agreement between DFA and rRT-PCR.
Values of Direct Fluorescent Antibody for diagnosis of Human Metapneumovirus (Hmpv)
| Parameter | Sensitivity | Specificity | PPV | NPV | Test efficiency |
|---|---|---|---|---|---|
| DFA | 50% | 100% | 100% | 95.8% | 96% |
5. Discussion
Acute respiratory tract infections have been major causes of morbidity and mortality, especially among the pediatric population (16). Even after extensive laboratory investigations, about half of ARTIs have no identifiable etiologic agent (6, 17). However, molecular techniques based on PCR technology can be employed to detect viruses that could be the culprits of some respiratory infections. In this scope, hMPV was described as a cause of respiratory infections, with a prevalence of 2 - 25% (18, 19).
Considering rRT-PCR as the gold standard test in the current study, hMPV was detected in 8% of the studied cases. This rate was consistent with that of Heikkinen et al. (20), who reported the incidence of hMPV infection as 7.6% in children < 2 years of age. Another study from Amman conducted by Schuster et al. (21) reported a detection rate of 8.6% among children < 2 years presenting with ARTI. Nonetheless, a higher detection rate was reported in some studies, where Arabpour et al. (22) reported that the overall frequency for hMPV infection among Iranian children < 2 years of age with ARTI was 54.4%. On the other hand, a lower detection rate was reported in other studies. A study conducted by Edwards et al. (23) detected hMPV in 5.1% out of 2806 hospitalized children under 24 months of age.
These variations could be affected by viral internal factors like the efficiency of viral replication, the virus ability to evade the host immune responses, the transmission route or environmental factors including geographical region, climate, seasonal fluctuation, yearly variations, and finally the genetic predilection of patients (22).
The present work revealed that there was no statistically significant difference between males and females regarding hMPV infection. This result was in agreement with Zou et al. (24) and Wang et al. (25).
In this study, the majority of hMPV-positive patients were detected during the winter months (11.5%) with the peak in February (16.7%). In line with our results, Ali et al. (26) from Pakistan reported February as the peak of hMPV activity (63%). However, a study from China stated that hMPV infections rather occurred throughout the year, with infection peaks during late winter and early spring (25).
Infants with underlying risk factors, particularly those with a history of prematurity or congenital heart disease, are at greater risk for unfavorable sequelae when infected with a respiratory virus (27). In this study, prematurity was found in one patient and co-morbidities were noticed in two patients, including cardiac disorder in one patient (25%) and allergy in the other one (25%). Similarly, Boivin et al. (28) found that 25% of children with hMPV infection had a cardiac disorder. Hence, the determination of risk factors for severe hMPV disease in young children can identify high-risk groups, who would benefit from preventive and therapeutic strategies (29).
In the present work, investigations revealed that infants with hMPV infection were more likely than those without the infection to have a higher lymphocytic count, possibly owing to the presence of specific viral diagnosis. Furthermore, chest X-ray in hMPV positive-patients was indifferent from hMPV negative cases. These findings assert that hMPV infections are moderate to severe in potency.
In our study, antibiotics and corticosteroids were by far the most frequently prescribed treatments followed by bronchodilators. Because at the time of consultation, no etiological pathogen had been detected, physicians continued treatment with antibiotics and corticosteroids to control potentially unidentified bacterial infections and to alleviate wheeze. This indicates that testing for hMPV in patients with ARTI may reduce unnecessary use of antibiotics and corticosteroids (30).
In this study, we found that one infant required Intensive Care Unit (ICU) admission and none of the patients with hMPV infection received mechanical ventilation. In some reports, none of the children infected with hMPV required ICU admission (28); however, others indicated that 15–25% of children required ICU care (31). Moreover, in the present work, there were no deaths associated with hMPV infection. This was also reported by Garcia-Garcia et al. (32) and Edwards et al. (23). Nonetheless, data revealed that hMPV may induce serious disease, and fatal outcomes have been reported (33, 34).
In our study, all hMPV positive cases were recovered from NPA, while ETA was negative by both DFA and rRT-PCR assays. This was in agreement with earlier studies, which recommended NPA and swab specimens for hMPV detection (10, 35).
In the present study, the mean delay between the onset of symptoms and sample collection was 3.24 ± 1.04. It was found that the longer the delay between symptom onset and sample collection, the more difficult it is to detect the causative agent. The majority of respiratory viruses are present in high titers in the respiratory tract in the first three days following symptom onset, whereas the viral nucleic acid may remain for a longer duration. Therefore, DFA loses sensitivity after the first three days post-onset of symptoms. In contrast to DFA, rRT-PCR represents a sensitive tool for virus detection even two weeks after symptom onset (36). The high sensitivity of rRT-PCR permits detection of viral nucleic acid even after the virus has disappeared, which renders it difficult to decide if the virus is the primary contributor to disease. Hence, nucleic acid detection results should be interpreted with caution (37).
The use of rapid tests for the diagnosis of hMPV infections allows implementation of proper infection control strategies, thus facilitating timely treatment (38). Good-quality smears and expertise in interpreting the results are required for reliable performance of the DFA assay (39).
In the present study, the two DFA-positive samples had a median CT value of 26.75, while the two rRT-PCR positive but DFA-negative samples had a median CT value of 28.75. These findings were consistent with the results of Landry et al. (14), who found that DFA-positive samples had a median CT value of 26.53, while rRT-PCR positive but DFA-negative samples had a median CT value of 36.18.
By considering the real-time assay as the gold standard for diagnosing hMPV in the current study, the sensitivity, specificity, PPV, NPV and test efficiency of DFA for the detection of hMPV were 50%, 100%, 100%, 95.8% and 96%, respectively. These were in agreement with a study conducted by Chang et al. (40), who stated that the DFA method showed sensitivity of 58.1%, specificity of 100%, PPV of 100%, NPV of 83% and test efficiency of 85.7% when compared with rRT-PCR assay.
In our study, the analytical specificity of DFA assay was very high (100%) and this yield ensures that DFA is a good negative test. Similar results were reported by different investigators: 94.1% in Italy (41), 97% in Japan (8), 100% in Canada (42) and 99% in Brazil (43). In contrast, the analytical sensitivity (50%) was found to be lower than studies carried out by Percivalle et al. (41) (73.9%), Ebihara et al. (8) (73.3%) and Vinh et al. (42) (95.2%). On the other hand, our sensitivity result was found to be higher than that by Wolf et al. (43), who reported a sensitivity of 39.5%. In addition, a study from Egypt reported by Zaki et al. (44) reported 100% sensitivity and 89% specificity of DFA when compared to RT-PCR assay and concluded that DFA can be securely used in hMPV screening tests.
The lower sensitivity of the DFA assay in our study in relation to PCR, could be explained by failure to detect hMPV viruses that have undergone minor changes in epitopes or due to the circulation of a new strain in one sample (viral load, 1 × 104 copies/mL) and the low number of virion particles, below the sensitivity of the DFA assay in the other sample (viral load, 500 copies/mL).
In the present study, DFA was found to be rapid and simple, requiring relatively little hands-on time in a clinical laboratory setting. However, it had poor performance. Many factors are related to this situation in a clinical laboratory setting. First, the MAb anti-hMPV needs to be specific to circulating strains to prevent non-specific background staining (14). Moreover, this technique has a lower sensitivity when compared with rRT-PCR (45). Also, the reader’s subjectivity, the need for specimens with appropriate number of cells and the impossibility of automation are other impediments associated with DFA (46). Therefore, combining the two methods, with the DFA assay as the first line, followed by RT-PCR for DFA-negative samples, may be the best approach to achieve prompt and sensitive detection of hMPV (17).
Most of hMPV-infected cases presented in the winter season, with pneumonia as the most common presentation of ARTIs in hMPV-infected patients. Therefore, routine testing for hMPV in infants with ARTI is imperative to avoid unnecessary antimicrobial therapy and to implement infection control precautions.
References
-
1.
Escobar C, Luchsinger V, de Oliveira DB, Durigon E, Chnaiderman J, Avendano LF. Genetic variability of human metapneumovirus isolated from Chilean children, 2003-2004. J Med Virol. 2009;81(2):340-4. [PubMed ID: 19107972]. https://doi.org/10.1002/jmv.21399.
-
2.
Gioula G, Chatzidimitriou D, Melidou A, Exindari M, Kyriazopoulou-Dalaina V. Contribution of human metapneumovirus to influenza-like infections in North Greece, 2005-2008. Euro Surveill. 2010;15(9). [PubMed ID: 20214868].
-
3.
Laguna-Torres VA, Sanchez-Largaespada JF, Lorenzana I, Forshey B, Aguilar P, Jimenez M, et al. Influenza and other respiratory viruses in three Central American countries. Influenza Other Respir Viruses. 2011;5(2):123-34. [PubMed ID: 21306576]. https://doi.org/10.1111/j.1750-2659.2010.00182.x.
-
4.
Kahn JS. Epidemiology of human metapneumovirus. Clin Microbiol Rev. 2006;19(3):546-57. [PubMed ID: 16847085]. https://doi.org/10.1128/CMR.00014-06.
-
5.
Koetz A, Nilsson P, Linden M, van der Hoek L, Ripa T. Detection of human coronavirus NL63, human metapneumovirus and respiratory syncytial virus in children with respiratory tract infections in south-west Sweden. Clin Microbiol Infect. 2006;12(11):1089-96. [PubMed ID: 17002608]. https://doi.org/10.1111/j.1469-0691.2006.01506.x.
-
6.
Sarasini A, Percivalle E, Rovida F, Campanini G, Genini E, Torsellini M, et al. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter--spring season. J Clin Virol. 2006;35(1):59-68. [PubMed ID: 16023411]. https://doi.org/10.1016/j.jcv.2005.05.010.
-
7.
Henrickson KJ. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23(1 Suppl):S6-10. [PubMed ID: 14730264]. https://doi.org/10.1097/01.inf.0000108187.63151.ea.
-
8.
Ebihara T, Endo R, Ma X, Ishiguro N, Kikuta H. Detection of human metapneumovirus antigens in nasopharyngeal secretions by an immunofluorescent-antibody test. J Clin Microbiol. 2005;43(3):1138-41. [PubMed ID: 15750074]. https://doi.org/10.1128/JCM.43.3.1138-1141.2005.
-
9.
Ebihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Hara M, et al. Human metapneumovirus infection in Japanese children. J Clin Microbiol. 2004;42(1):126-32. [PubMed ID: 14715742].
-
10.
van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J. 2004;23(1 Suppl):S25-32. [PubMed ID: 14730267]. https://doi.org/10.1097/01.inf.0000108190.09824.e8.
-
11.
World Health Organization Hand book integrated management of childhood illnesses. Geneva: World Health Organization; 2005. Available from: http://www.who.int/maternal_child_adolescent.
-
12.
Heikkinen T, Marttila J, Salmi AA, Ruuskanen O. Nasal swab versus nasopharyngeal aspirate for isolation of respiratory viruses. J Clin Microbiol. 2002;40(11):4337-9. [PubMed ID: 12409425].
-
13.
Zhang C, Du LN, Zhang ZY, Qin X, Yang X, Liu P, et al. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Southwest China. J Clin Microbiol. 2012;50(8):2714-9. [PubMed ID: 22692746]. https://doi.org/10.1128/JCM.00809-12.
-
14.
Landry ML, Cohen S, Ferguson D. Prospective study of human metapneumovirus detection in clinical samples by use of light diagnostics direct immunofluorescence reagent and real-time PCR. J Clin Microbiol. 2008;46(3):1098-100. [PubMed ID: 18184854]. https://doi.org/10.1128/JCM.01926-07.
-
15.
Al-Turab M, Chehadeh W, Al-Mulla F, Al-Nakib W. Evaluation of the PrimerDesign genesig real-time reverse transcription-polymerase chain reaction assay and the INFINITI(R) Respiratory Viral Panel Plus assay for the detection of human metapneumovirus in Kuwait. Diagn Microbiol Infect Dis. 2012;72(4):358-62. [PubMed ID: 22300956]. https://doi.org/10.1016/j.diagmicrobio.2012.01.001.
-
16.
Zar HJ. Respiratory infections in children in developing countries. Pediatr Ann. 2002;31(2):133-8. [PubMed ID: 11862724].
-
17.
Gerna G, Campanini G, Rovida F, Sarasini A, Lilleri D, Paolucci S, et al. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter-spring seasons. Brief report. Arch Virol. 2005;150(11):2365-75. [PubMed ID: 15986171]. https://doi.org/10.1007/s00705-005-0581-2.
-
18.
Carr MJ, McCormack GP, Crowley B. Human metapneumovirus-associated respiratory tract infections in the Republic of Ireland during the influenza season of 2003-2004. Clin Microbiol Infect. 2005;11(5):366-71. [PubMed ID: 15819862]. https://doi.org/10.1111/j.1469-0691.2005.01129.x.
-
19.
Williams JV. Human Metapneumovirus: An Important Cause of Respiratory Disease in Children and Adults. Curr Infect Dis Rep. 2005;7(3):204-10. [PubMed ID: 15847723].
-
20.
Heikkinen T, Osterback R, Peltola V, Jartti T, Vainionpaa R. Human metapneumovirus infections in children. Emerg Infect Dis. 2008;14(1):101-6. [PubMed ID: 18258088]. https://doi.org/10.3201/eid1401.070251.
-
21.
Schuster JE, Khuri-Bulos N, Faouri S, Shehabi A, Johnson M, Wang L, et al. Human Metapneumovirus Infection in Jordanian Children: Epidemiology and Risk Factors for Severe Disease. Pediatr Infect Dis J. 2015;34(12):1335-41. [PubMed ID: 26372450]. https://doi.org/10.1097/INF.0000000000000892.
-
22.
Arabpour M, Samarbafzadeh AR, Makvandi M, Shamsizadeh A, Percivalle E, Englud J, et al. The highest prevalence of human metapneumovirus in Ahwaz children accompanied by acute respiratory infections. Indian J Med Microbiol. 2008;26(2):123-6. [PubMed ID: 18445946].
-
23.
Edwards KM, Zhu Y, Griffin MR, Weinberg GA, Hall CB, Szilagyi PG, et al. Burden of human metapneumovirus infection in young children. N Engl J Med. 2013;368(7):633-43. [PubMed ID: 23406028]. https://doi.org/10.1056/NEJMoa1204630.
-
24.
Zou LR, Mo YL, Wu D, Fang L, Li H, Chen QX, et al. [Investigation of human metapneumovirus in children with acute respiratory tract infections in Guangzhou areas]. Zhonghua Yu Fang Yi Xue Za Zhi. 2009;43(4):314-8. [PubMed ID: 19534954].
-
25.
Wang Y, Chen Z, Yan YD, Guo H, Chu C, Liu J, et al. Seasonal distribution and epidemiological characteristics of human metapneumovirus infections in pediatric inpatients in Southeast China. Arch Virol. 2013;158(2):417-24. [PubMed ID: 23074040]. https://doi.org/10.1007/s00705-012-1492-7.
-
26.
Ali A, Khowaja AR, Bashir MZ, Aziz F, Mustafa S, Zaidi A. Role of human metapneumovirus, influenza A virus and respiratory syncytial virus in causing WHO-defined severe pneumonia in children in a developing country. PLoS One. 2013;8(9):74756. [PubMed ID: 24058625]. https://doi.org/10.1371/journal.pone.0074756.
-
27.
Durigon GS, Oliveira DB, Felicio MC, Finelli C, Pereira MF, Storni JG, et al. Poor outcome of acute respiratory infection in young children with underlying health condition in Brazil. Int J Infect Dis. 2015;34:3-7. [PubMed ID: 25747778]. https://doi.org/10.1016/j.ijid.2015.03.003.
-
28.
Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, et al. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis. 2003;9(6):634-40. [PubMed ID: 12781001]. https://doi.org/10.3201/eid0906.030017.
-
29.
Roussy JF, Carbonneau J, Ouakki M, Papenburg J, Hamelin ME, De Serres G, et al. Human metapneumovirus viral load is an important risk factor for disease severity in young children. J Clin Virol. 2014;60(2):133-40. [PubMed ID: 24686044]. https://doi.org/10.1016/j.jcv.2014.03.001.
-
30.
Yahia S, Kandeel AY, Hammad E, El-Gilany AH. Human metapneumovirus (hMPV) in acute respiratory infection: a clinic-based study in Egypt. Indian J Pediatr. 2012;79(10):1323-7. [PubMed ID: 22294269]. https://doi.org/10.1007/s12098-011-0677-5.
-
31.
Hamelin ME, Abed Y, Boivin G. Human metapneumovirus: a new player among respiratory viruses. Clin Infect Dis. 2004;38(7):983-90. [PubMed ID: 15034830]. https://doi.org/10.1086/382536.
-
32.
Garcia-Garcia ML, Calvo C, Martin F, Perez-Brena P, Acosta B, Casas I. Human metapneumovirus infections in hospitalised infants in Spain. Arch Dis Child. 2006;91(4):290-5. [PubMed ID: 16399780]. https://doi.org/10.1136/adc.2005.082388.
-
33.
Noyola DE, Alpuche-Solis AG, Herrera-Diaz A, Soria-Guerra RE, Sanchez-Alvarado J, Lopez-Revilla R. Human metapneumovirus infections in Mexico: epidemiological and clinical characteristics. J Med Microbiol. 2005;54(Pt 10):969-74. [PubMed ID: 16157552]. https://doi.org/10.1099/jmm.0.46052-0.
-
34.
Morrow BM, Hatherill M, Smuts HE, Yeats J, Pitcher R, Argent AC. Clinical course of hospitalised children infected with human metapneumovirus and respiratory syncytial virus. J Paediatr Child Health. 2006;42(4):174-8. [PubMed ID: 16630317]. https://doi.org/10.1111/j.1440-1754.2006.00825.x.
-
35.
van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7(6):719-24. [PubMed ID: 11385510]. https://doi.org/10.1038/89098.
-
36.
Shafik CF, Mohareb EW, Yassin AS, Amin MA, El Kholy A, El-Karaksy H, et al. Viral etiologies of lower respiratory tract infections among Egyptian children under five years of age. BMC Infect Dis. 2012;12:350. [PubMed ID: 23237512]. https://doi.org/10.1186/1471-2334-12-350.
-
37.
Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol. 2004;72(4):695-9. [PubMed ID: 14981776]. https://doi.org/10.1002/jmv.20027.
-
38.
Macfarlane P, Denham J, Assous J, Hughes C. RSV testing in bronchiolitis: which nasal sampling method is best? Arch Dis Child. 2005;90(6):634-5. [PubMed ID: 15908632]. https://doi.org/10.1136/adc.2004.065144.
-
39.
Rovida F, Percivalle E, Zavattoni M, Torsellini M, Sarasini A, Campanini G, et al. Monoclonal antibodies versus reverse transcription-PCR for detection of respiratory viruses in a patient population with respiratory tract infections admitted to hospital. J Med Virol. 2005;75(2):336-47. [PubMed ID: 15602736]. https://doi.org/10.1002/jmv.20276.
-
40.
Chang YF, Tsao KC, Liu YC, Chen YC, Yu PC, Huang YC, et al. Diagnosis of human metapneumovirus in patients hospitalized with acute lower respiratory tract infection using a metal-enhanced fluorescence technique. J Virol Methods. 2015;213:151-6. [PubMed ID: 25522922]. https://doi.org/10.1016/j.jviromet.2014.12.005.
-
41.
Percivalle E, Sarasini A, Visai L, Revello MG, Gerna G. Rapid detection of human metapneumovirus strains in nasopharyngeal aspirates and shell vial cultures by monoclonal antibodies. J Clin Microbiol. 2005;43(7):3443-6. [PubMed ID: 16000473]. https://doi.org/10.1128/JCM.43.7.3443-3446.2005.
-
42.
Vinh DC, Newby D, Charest H, McDonald J. Evaluation of a commercial direct fluorescent-antibody assay for human metapneumovirus in respiratory specimens. J Clin Microbiol. 2008;46(5):1840-1. [PubMed ID: 18367577]. https://doi.org/10.1128/JCM.01554-07.
-
43.
Wolf JM, Gregianini TS, Seadi CM, Tumioto GL, Dambros BP, Lehmann FK, et al. Performance of direct immunofluorescence assay for the detection of human metapneumovirus under clinical laboratory settings. Rev Soc Bras Med Trop. 2015;48(6):762-4. [PubMed ID: 26676503]. https://doi.org/10.1590/0037-8682-0107-2015.
-
44.
Zaki WK, S. Fathi M, Ismail R. Detection of Human Metapneumo Virus among Infants with Bronchiolitis. International J Virol. 2014;10(1):37-45. https://doi.org/10.3923/ijv.2014.37.45.
-
45.
Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716-47. [PubMed ID: 18854489]. https://doi.org/10.1128/CMR.00037-07.
-
46.
Kanashiro TM, Vilas Boas LS, Thomaz AM, Tozetto-Mendoza TR, Setsuko M, Machado CM. Identification of respiratory virus in infants with congenital heart disease by comparison of different methods. Rev Inst Med Trop Sao Paulo. 2011;53(5):241-6. [PubMed ID: 22012448].