Abstract
Background:
The presence of carbapenemase-producing Acinetobacter baumannii has become a growing concern in patients who are hospitalized in burns centers.Objectives:
The aims of this study were to determine the antimicrobial susceptibility patterns and prevalence of blaOXA carbapenemases, as well as to detect the presence of ISAba1, in A. baumannii strains carrying OXA genes obtained from burns patients at Shahid Motahari hospital, Tehran, Iran.Methods:
From August 2013 to March 2014, 100 clinical A. baumannii isolates were collected from patients who were admitted to the burns ward at Shahid Motahari hospital. Antimicrobial susceptibility was determined using a disc diffusion test. PCR, sequencing, and multiplex PCR were used for the detection of blaOXA-23-like, blaOXA-51-like, blaOXA-24-like, and blaOXA-58-like genes, which were then sequenced. The ISAba1 gene was detected, and PCR was performed to detect the presence of ISAba1/blaOXA-51-like and ISAba1/blaOXA-23-like genes.Results:
The results showed that 93% of the strains were multi-drug resistant, while 82% of them were extensively drug resistant. Additionally, all the strains carried blaOXA-23-like and blaOXA-51-like genes, while 74% and 0% of the strains harbored blaOXA-24-like and blaOXA-58 genes, respectively. ISAba1 was detected in all the strains except for one. The co-existence of ISAba1/blaOXA-51-like genes and ISAba1/blaOXA-23-like genes was detected in 65% and 80% of strains, respectively.Conclusions:
The results of this study indicate that the emergence of OXA-type carbapenemases in A. baumannii causing nosocomial infections in burns patients could be of importance for hospital infection control systems in Iran.Keywords
Carbapenemase Isaba1 Multi-Drug Resistance Extensively Drug Resistance Burn Acinetobacter baumannii
1. Background
Burns patients are at risk of acquiring infection due to their damaged skin and impaired immune system (1). Acinetobacter baumannii is an opportunistic pathogen that appears to have become one of the most important causes of nosocomial infections in hospitalized patients, particularly those in burns units, in recent years (2-5). During the past decade, this pathogen has been reported to be the second most common cause of nosocomial infections in burns patients (3, 6-8). The nosocomial infection strains of A. baumannii are multi-drug resistant (MDR) and extensively drug resistant (XDR) due to increased rates of resistance to the most commonly available antibiotics, including carbapenemas, which is the drug of choice for treating infection with A. baumannii (9-12). One of the most common mechanisms in carbapenem resistance is the production of carbapenem-hydrolyzing β-lactamase enzymes (carbapenemases) by these strains (2, 10, 11, 13-15). Two molecular classes of carbapenemases, namely Ambler class B (metallo-β-lactamase) and Ambler class D (oxacillinase), have been identified, with class D being the most prevalent enzyme among strains of A. baumannii (2, 6, 13, 16, 17). The genetic analysis of OXA-type enzymes (encoded by blaOXA genes) has categorized them into eight distinctive subgroups. Four of them OXA-51-like, OXA-23-like, OXA-24-like, and OXA-58-like have been found in A. baumannii (2, 11, 14, 18, 19). According to reports from different countries, blaOXA-51-like genes are intrinsically harbored by A. baumannii isolates, although their expression varies according to the presence of an insertion sequence such as ISAba1 on the upstream of the gene (11, 20, 21). In addition, other OXA carbapenemase genes that are not part of the normal genome of the species can inactivate carbapenems, albeit less efficiently, and their presence or activation by ISAba1 is correlated with resistance (21-23).
2. Objectives
The aims of this study were to survey the drug resistance patterns of A. baumannii strains isolated from burns patients with nosocomial infections at Shahid Motahari hospital in Tehran, Iran, as well as to identify the genes encoding the ISAba1 element and the four subgroups of OXA-type carbapenemases among these isolates. The study also sought to determine the co-existence of ISAba1/blaOXA-51-like genes and ISAba1/blaOXA-23-like genes.
3. Methods
3.1. Bacterial Strains
This study was conducted at Shahid Motahari hospital, a burns center in Tehran, Iran. One hundred samples of A. baumannii isolated from patients’ burn wounds were collected between August 2013 and March 2014. Prior to sampling, the wounds were washed using physiological serum. The samples were transferred to Stuart media, cultured on blood agar and MacConkey agar, and then incubated at 37°C for 24 hours. Conventional biochemical tests such as oxidase, triple sugar iron (TSI), sulfide indole motility (SIM), and growing at 44°C were used for the primary identification of A. baumannii strains. The identification of the isolates was confirmed by blaOXA-51-like gene detection (5).
3.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility testing was performed using the disk agar diffusion method on Mueller-Hinton agar according to Clinical and Laboratory Standards Institute (CLSI) guidelines (24). Disks containing cefepime (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), ceftriaxon (30 μg), cefotaxime (30 μg), imipenem (10 μg), meropenem (10 μg), piperacillin-tazobactam (100/10 μg), gentamicin (10 μg), amikacin (30 μg), tetracycline (30 μg), trimethoprim-sulfamethoxazole (5 μg), and piperacillin (100 μg) were placed on a plate. Then, the samples were incubated at 37°C for 24 hours. The minimum inhibitory concentrations (MICs) of the imipenem, meropenem, ceftazidime, ciprofloxacin, and colistin were determined according to the microdilution broth method. The standard antibiotic disks and powders used in this study were obtained from Mast Diagnostics (Mast Group Ltd., Bootle, UK). P. aeruginosa ATCC 27853 was used as the control strain in each susceptibility test.
3.3. PCR Amplification for the blaOXA Genes and ISAba1 Element
DNA was extracted from the strains using a PrimePrep Genomic DNA Isolation Kit (Cat. No. K-3000, GENETBIO Inc., Daejeon, South Korea) according to the manufacturer’s instructions. The 25 µL PCR mixture contained 2 µL of bacterial DNA, 10 pM of each primer, 250 µM of each dNTP, 1.5 mM of MgCl2, 10 Mm of Tris-HCL, 30 Mm of KCL, and 1 U of Taq DNA polymerase (Cat. No. K-2012, Bioneer Company, Korea). The reactions were performed in a thermal cycler (Mastercycler Gradient, Eppendorf, Hamburg, Germany). To amplify the genes encoding the carbapenemases, PCR assays were run for the blaOXA-23-like, blaOXA-51-like, blaOXA-24-like, and blaOXA-58-like genes and the ISAba1 element. In addition, a set of multiplex PCRs for the blaOXA-23-like, blaOXA-51-like, and blaOXA-24-like genes were designed using an internal positive control for each gene. The primers used to amplify these genes are listed in Table 1, while the PCR system conditions for the amplification of each gene are presented in Table 2. Furthermore, PCRs were performed to detect the ISAba1/blaOXA-51-like and ISAba1/blaOXA-23-like sequences using a combination of the ISAba1 forward primers and the blaOXA-51-like and blaOXA-23-like reverse primers (Table 1). The PCR products were analyzed using 1.5% agarose gel electrophoresis with ethidium bromide staining.
Sequence of Primers
| Primer | Sequence (5’ to 3’) | Target | References |
|---|---|---|---|
| OXA-23-likeF | GATCGGATTGGAGAACCAGA | blaOXA-23-like | (25) |
| OXA-23-likeR | ATTTCTGACCGCATTTCCAT | ||
| OXA-24-likeF | GGTTAGTTGGCCCCCTTAAA | blaOXA-24-like | (25) |
| OXA-24-likeR | AGTTGAGCGAAAAGGGGATT | ||
| OXA-58-likeF | AAGTATTGGGGCTTGTGCTG | blaOXA-58-like | (25) |
| OXA-58-likeR | CCCCTCTGCGCTCTACATAC | ||
| ISAba1F | CACGAATGCAGAAGTTG | ISAba1 | (26) |
| ISAba1R | CGACGAATACTATGACAC | ||
| ISAba1F1 | AGGCTATAAAGCGTTGA | ISAba1/blaOXA-51-like | (27) |
| OXA-51-likeR1 | CTTCTGTGGTGGTTGC | ||
| ISAba1F2 | AACGATTGCGAGCATC | ISAba1/blaOXA-23-like | (27) |
| OXA-23-likeR2 | GTCAACCAGCCCACTT |
PCR Conditions for the bla Genes Amplification
| Factor | Temperature (°C) | Time | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaOXA-23 | Blaoxa-51 | Blaoxa-24 | Blaoxa-58 | ISAba1 | ISAba1/blaOXA-23 | ISAba1/blaoxa-51 | ||||||||
| Initial denaturation | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 5 min | 5 min | 5 min | 7 min | 5 min | 5 min | 5 min |
| Denaturation | 94 | 94 | 94 | 94 | 94 | 94 | 94 | 30 s | 30 s | 30 s | 1 min | 45 s | 45 s | 45 s |
| Annealing | 53 | 53 | 54 | 53 | 41 | 54 | 52 | 45 s | 45 s | 45 s | 1 min | 1 min | 1 min | 1 min |
| Extension | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 45 s | 45 s | 45 s | 1 min | 45 s | 45 s | 45 s |
| Final extension | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 5 min | 5 min | 5 min | 5 min | 5 min | 5 min | 5 min |
| Cycle | 36 | 36 | 36 | 36 | 36 | 36 | 36 | |||||||
3.4. Sequencing Technique
The PCR products were purified using a PCR purification kit (Bioneer Co., Korea). The direct sequencing of the amplicons was performed by the Bioneer Company (Korea). The nucleotide sequences were analyzed using FinchTV software and BLAST in NCBI.
3.5. Statistical Analysis
MINITAB 16 software was used for all the statistical analyses in this study. This proposal was accepted by Medical Ethics in Shahid Beheshti University of Medical Sciences, (IR.SBMU.RAM.REC.1394.213).
4. Results
One hundred strains of A. baumannii were isolated from burns patients admitted to Shahid Motahari hospital. Of the 100 strains obtained from patients’ burn wounds, 26 strains were isolated from female patients (26%) and 74 strains from males (74%). The patients were aged from 1- to 90-years-old (Table 3).
Distribution of Patients Infected with A. baumannii by Age
| Age | Number of Patients (%) |
|---|---|
| 1 - 15 | 5 (5) |
| 16 - 30 | 5 (5) |
| 31 - 45 | 31 (31) |
| 46 - 60 | 36 (36) |
| 61 - 75 | 17 (17) |
| 76 - 90 | 6 (6) |
| Total | 100 (100) |
4.1. Antimicrobial Susceptibility
Based on the antimicrobial susceptibility testing, high rates of resistance to cefotaxime (100%), ceftriaxone (100%), ceftazidime (100%), meropenem (98%), imipenem (98%), gentamicin (93%), amikacin (90%), ciprofloxacin (100%), cefepime (100%), piperacillin/tazobactam (100%), tetracycline (82%), piperacillin (99%), and trimethoprim/sulfamethoxazole (95%) were observed (Table 4). Some 93% of strains were resistant to all the tested antibiotics, while 98% of them showed resistance to all the tested carbapenems. The MIC values of imipenem, meropenem, colistin, ceftazidime, and ciprofloxacin in relation to the A. baumannii isolates are shown in Table 5. Colistin was found to be the most effective drug against A. baumannii strains, with a 100% susceptibility rate being reported. In this study, all the isolates that had OXA-type genes were carbapenem resistant. The results showed that 93% of the strains were MDR, while 82% of them were XDR.
Antimicrobial Susceptibility Pattern of A. baumannii Isolates According to the Disc Diffusion Method
| Antimicrobials | Susceptible No. (%) | Intermediate No. (%) | Resistant No. (%) |
|---|---|---|---|
| Imipenem | 0 | 2 (2) | 98 (98) |
| Meropenem | 1 (1) | 1 (1) | 98 (98) |
| Ceftriaxon | 0 | 0 | 100 (100) |
| Cefotaxime | 0 | 0 | 100 (100) |
| Ceftazidime | 0 | 0 | 100 (100) |
| Cefepime | 0 | 0 | 100 (100) |
| Ciprofloxacin | 0 | 0 | 100 (100) |
| Trimetoprim-sulfametoxazole | 4 (4) | 2 (2) | 94 (94) |
| Amikacin | 5 (5) | 5 (5) | 90 (90) |
| Tetracycline | 10 (10) | 8 (8) | 82 (82) |
| Gentamycin | 5 (5) | 2 (2) | 93 (93) |
| Piperacillin | 0 | 0 | 100 (100) |
| Piperacillin Tazobactam | 0 | 1 (1) | 99 (99) |
The MIC Values of the Antibiotics Against A. baumannii
| Antibiotic | Susceptible No. (%) | Intermediate No. (%) | Resistant No. (%) |
|---|---|---|---|
| Imipenem | 3 (3) | 3 (3) | 94 (94) |
| Meropenem | 6 (6) | 5 (5) | 89 (89) |
| Ceftazidime | 0 | 0 | 100 (100) |
| Ciprofloxacin | 0 | 2 (2) | 98 (98) |
| Colistin | 100 (100) | 0 | 0 |
4.2. Carbapenemases of A. baumannii
The blaOXA-51-like gene was identified in all the A. baumannii strains (100%) by PCR and multiplex PCR. While it was not necessarily related to the increased resistance among these isolates, the gene was associated with the insertion of ISAba1, overexpression, and reduced susceptibility to carbapenems. Although the prevalence of blaOXA-23-like and blaOXA-24-like genes was 100% and 74%, respectively, the blaOXA-58-like gene was not detected in any of these isolates (Figure 1). The ISAba1 element was identified in all the isolates except for one (99%). The co-existence of ISAba1/blaOXA-51-like genes and ISAba1/blaOXA-23-like genes was detected in 65% and 80% of strains, respectively.
Multiplex PCR Amplification of the OXA-24, OXA-23, and OXA-51 Genes of A. baumannii Isolates
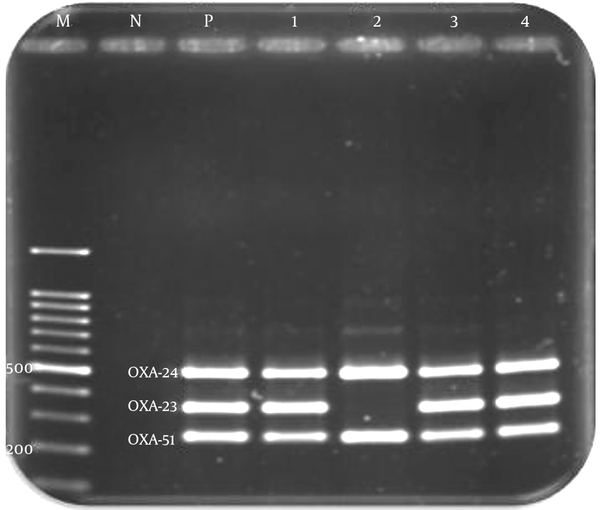
4.3. Sequencing
The sequencing of the PCR products showed conserved regions for the blaOXA-23-like, blaOXA-51-like, blaOXA-24-like, and ISAba1 genes, which were confirmed with the basic local alignment search tool (BLAST) of the national center for biotechnology information (NCBI). The nucleotide sequence data reported in this paper have been submitted to the GenBank sequence database, and accession numbers KT313639 and KT313638 have been assigned for the OXA-23 and OXA-24 encoding genes, respectively.
5. Discussion
According to previous studies, A. baumannii is an emerging nosocomial pathogen in burns patients due to the potential for acquiring resistance to multiple antimicrobial agents, which often leads to mortality and morbidity in infected patients (28, 29). A broad spectrum of antimicrobial resistance, including carbapenems, is an important challenge for Iranian burns patients infected with A. baumannii, since treating them is difficult due to the limited number of antibiotics that remain effective against OXA-type producing A. baumannii strains such as polymyxin B and colistin. Hence, the emergence of resistance to these agents and the neurological risks arising from them should be considered (6, 10).
The antibiotic resistance of A. baumannii strains is increasing day by day, which leads to the emergence of MDR and XDR strains because of the indiscriminate use of a broad spectrum of antimicrobial agents and the high ability of these isolates to acquire multiple resistant genes. The results of this study showed that 93% of the strains isolated from burns patients were MDR, while 82% of them were XDR. The results of several prior studies have shown that antimicrobial resistance is increasing worldwide (30, 31). Further, an increasing trend of carbapenem resistance from 2009 until the present day is visible (30, 32).
OXA-type carbapenemases are the dominant enzymes involved in antibiotic-resistant A. baumannii strains, especially the OXA-23-like genes, which have been reported worldwide (27, 33). OXA-51-like encoded genes naturally occur in A. baumannii isolates, and they have also been identified worldwide (34, 35). In our study, all the strains had intrinsically blaOXA-51-like genes, and the identification of the isolates as A. baumannii has been confirmed. The presence of blaOXA-23-like genes was observed in all the strains. In 2013, OXA-23-like encoded genes were detected in 94% of isolated A. baumannii in China (36). Additionally, the results of a study conducted in Iran in 2012 indicated that blaOXA-23-like genes were detected in all the strains (37), which is consistent with our results.
The OXA-24 group has been reported in Portugal, Spain, Belgium, France, and the United States (38). In addition, strains producing blaOXA-58-like genes were found in isolates recovered from Italy, Belgium, France, Greece, the United States, and Argentina (34). Although blaOXA-24-like genes were detected in 74% of the A. baumannii isolates in our study, this rate was lower than that reported in previous studies (34, 37, 39). The differing results may be due to the different geographical locations, different clinical samples, and various antibiotype patterns in the different studies.
The absence of OXA-58-like encoded genes in all the isolates in our study is consistent with the findings of similar studies (34, 37, 40). The important thing is that an ISAba1 element was detected in all the A. baumannii strains. The presence of the ISAba1 insertion upstream of the OXA-51 and OXA-23 was seen in 63% and 80% of the isolates, respectively. This co-existence has been shown to confer high levels of carbapenem resistance (20). In 2012, the incidence of co-existence with ISAba1 was 80% and 66% for blaOXA-51 and blaOXA-23, respectively, in Turkey (32). Further, the ISAab1 element was detected in the upstream of 85% of blaOXA-51 genes and 80% of blaOXA-23 genes in Egypt in 2012 (39). All these results demonstrate a high level of similarity to the findings of our study, and they emphasize the presence of ISAba1 upstream of blaOXA-23-like genes in instances of resistance enhancement.
The results of this study indicate the emergence of OXA-type carbapenemases in A. baumannii causing nosocomial infections in burns patients, which could be of importance for hospital infection control systems in Iran. Additionally, A. baumannii exhibiting the co-existence of OXA genes and ISAba1 are extremely prevalent in Iran, which may cause serious problems during the treatment of A. baumannii infections using antibiotics
Acknowledgements
References
-
1.
Hakemi Vala M, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Sattarzadeh Tabrizi M, et al. Detection of Ambler class A, B and D ss-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters. 2014;27(1):8-13. [PubMed ID: 25249841].
-
2.
Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471-84. [PubMed ID: 17646423]. https://doi.org/10.1128/AAC.01464-06.
-
3.
Peymani A, Nahaei MR, Farajnia S, Hasani A, Mirsalehian A, Sohrabi N, et al. High prevalence of metallo-beta-lactamase-producing acinetobacter baumannii in a teaching hospital in Tabriz, Iran. Jpn J Infect Dis. 2011;64(1):69-71. [PubMed ID: 21266761].
-
4.
Nordmann P. Acinetobacter baumannii, the nosocomial pathogen par excellence [in French]. Pathol Biol (Paris). 2004;52(6):301-3. [PubMed ID: 15261370]. https://doi.org/10.1016/j.patbio.2004.03.001.
-
5.
Noori M, Karimi A, Fallah F, Hashemi A, Alimehr S, Goudarzi H, et al. High Prevalence of Metallo-beta-lactamase Producing Acinetobacter baumannii Isolated From Two Hospitals of Tehran, Iran. Arch Pediatr Infect Dis. 2014;2(3). https://doi.org/10.5812/pedinfect.15439.
-
6.
Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(4):274-8. [PubMed ID: 18653968].
-
7.
Alaghehbandan R, Azimi L, Rastegar Lari A. Nosocomial infections among burn patients in Teheran, Iran: a decade later. Ann Burns Fire Disasters. 2012;25(1):3-7. [PubMed ID: 23012608].
-
8.
Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977-2000. Infect Control Hosp Epidemiol. 2003;24(4):284-95. [PubMed ID: 12725359]. https://doi.org/10.1086/502205.
-
9.
Garcia-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, Jimenez-Jimenez FJ, Perez-Paredes C, Barrero-Almodovar AE, et al. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin Infect Dis. 2001;33(7):939-46. [PubMed ID: 11528563]. https://doi.org/10.1086/322584.
-
10.
Azimi L, Talebi M, Pourshafie MR, Owlia P, Rastegar Lari A. Characterization of Carbapenemases in Extensively Drug Resistance Acinetobacter baumannii in a Burn Care Center in Iran. Int J Mol Cell Med. 2015;4(1):46-53. [PubMed ID: 25815282].
-
11.
Brown S, Young HK, Amyes SG. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect. 2005;11(1):15-23. [PubMed ID: 15649299]. https://doi.org/10.1111/j.1469-0691.2004.01016.x.
-
12.
Fallah F, Noori M, Hashemi A, Goudarzi H, Karimi A, Erfanimanesh S, et al. Prevalence of bla NDM, bla PER, bla VEB, bla IMP, and bla VIM Genes among Acinetobacter baumannii Isolated from Two Hospitals of Tehran, Iran. Scientifica (Cairo). 2014;2014:245162. [PubMed ID: 25133013]. https://doi.org/10.1155/2014/245162.
-
13.
Naas T, Nordmann P. OXA-type beta-lactamases. Curr Pharm Des. 1999;5(11):865-79. [PubMed ID: 10539993].
-
14.
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-6. [PubMed ID: 16891520]. https://doi.org/10.1128/JCM.01021-06.
-
15.
Fallah F, Taherpour A, Borhan RS, Hashemi A, Habibi M, Sajadi Nia R. Evaluation of Zataria MultiFlora Boiss and Carum copticum antibacterial activity on IMP-type metallo-beta-lactamase-producing Pseudomonas aeruginosa. Ann Burns Fire Disasters. 2013;26(4):193-8. [PubMed ID: 24799849].
-
16.
Roodsari MR, Fallah F, Taherpour A, Vala MH, Hashemi A. Carbapenem-resistant bacteria and laboratory detection methods. Arch Pediatr Infect Dis. 2014;2(1):188-91.
-
17.
Goudarzi H, Taherpour A, Fallah F, Pourkaveh B, Erfanimanesh S, Hashemi A. Laboratory Detection of Carbapenemases in Gram-Negative Bacteria. Arch Clin Infect Dis. 2016;In Press(In Press). https://doi.org/10.5812/archcid.32816.
-
18.
Paton R, Miles RS, Hood J, Amyes SG, Miles RS, Amyes SG. ARI 1: beta-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2(2):81-7. [PubMed ID: 18611526].
-
19.
Fallah F, Taherpour A, Vala MH, Hashemi A. Global Spread of New Delhi mettallo-beta-lactamase-1 (NDM-1). Arch Clin Infect Dis. 2012;6(4):171-7.
-
20.
Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett. 2006;258(1):72-7. [PubMed ID: 16630258]. https://doi.org/10.1111/j.1574-6968.2006.00195.x.
-
21.
Heritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother. 2005;49(10):4174-9. [PubMed ID: 16189095]. https://doi.org/10.1128/AAC.49.10.4174-4179.2005.
-
22.
Villegas MV, Kattan JN, Correa A, Lolans K, Guzman AM, Woodford N, et al. Dissemination of Acinetobacter baumannii clones with OXA-23 Carbapenemase in Colombian hospitals. Antimicrob Agents Chemother. 2007;51(6):2001-4. [PubMed ID: 17403994]. https://doi.org/10.1128/AAC.00226-07.
-
23.
Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006;57(1):1-3. [PubMed ID: 16332731]. https://doi.org/10.1093/jac/dki425.
-
24.
Performance standards for antimicrobial susceptibility testing; Twenty-second informational supplement. California: CLSI; 2012, [cited Jan]. Available from: antimicrobianos.com.ar/ATB/wp-content/uploads/2012/11/M100S22E.pdf.
-
25.
Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351-3. [PubMed ID: 16564159]. https://doi.org/10.1016/j.ijantimicag.2006.01.004.
-
26.
Hong SB, Shin KS, Ha J, Han K. Co-existence of blaOXA-23 and armA in multidrug-resistant Acinetobacter baumannii isolated from a hospital in South Korea. J Med Microbiol. 2013;62(Pt 6):836-44. [PubMed ID: 23518656]. https://doi.org/10.1099/jmm.0.055384-0.
-
27.
Zhou H, Pi BR, Yang Q, Yu YS, Chen YG, Li LJ, et al. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1 blaOXA-23 genes in a Chinese hospital. J Med Microbiol. 2007;56(8):1076-80. [PubMed ID: 17644715]. https://doi.org/10.1099/jmm.0.47206-0.
-
28.
Taherikalani M, Fatolahzadeh B, Emaneini M, Soroush S, Feizabadi MM. Distribution of different carbapenem resistant clones of Acinetobacter baumannii in Tehran hospitals. New Microbiol. 2009;32(3):265-71. [PubMed ID: 19845108].
-
29.
Ku WW, Kung CH, Lee CH, Tseng CP, Wu PF, Kuo SC, et al. Evolution of carbapenem resistance in Acinetobacter baumannii: an 18-year longitudinal study from a medical center in northern Taiwan. J Microbiol Immunol Infect. 2015;48(1):57-64. [PubMed ID: 24064289]. https://doi.org/10.1016/j.jmii.2013.07.005.
-
30.
Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;53(9):3628-34. [PubMed ID: 19528270]. https://doi.org/10.1128/AAC.00284-09.
-
31.
Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: a descriptive study in a city hospital. BMC Infect Dis. 2010;10:196. [PubMed ID: 20609238]. https://doi.org/10.1186/1471-2334-10-196.
-
32.
Cicek AC, Saral A, Iraz M, Ceylan A, Duzgun AO, Peleg AY, et al. OXA- and GES-type beta-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish University Hospital. Clin Microbiol Infect. 2014;20(5):410-5. [PubMed ID: 23957892]. https://doi.org/10.1111/1469-0691.12338.
-
33.
Poirel L, Nordmann P. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob Agents Chemother. 2006;50(4):1442-8. [PubMed ID: 16569863]. https://doi.org/10.1128/AAC.50.4.1442-1448.2006.
-
34.
Nowak P, Paluchowska P, Budak A. Distribution of blaOXA genes among carbapenem-resistant Acinetobacter baumannii nosocomial strains in Poland. New Microbiol. 2012;35(3):317-25. [PubMed ID: 22842601].
-
35.
Wroblewska MM, Towner KJ, Marchel H, Luczak M. Emergence and spread of carbapenem-resistant strains of Acinetobacter baumannii in a tertiary-care hospital in Poland. Clin Microbiol Infect. 2007;13(5):490-6. [PubMed ID: 17331123]. https://doi.org/10.1111/j.1469-0691.2007.01694.x.
-
36.
Gao J, Zhao X, Bao Y, Ma R, Zhou Y, Li X, et al. Antibiotic resistance and OXA-type carbapenemases-encoding genes in airborne Acinetobacter baumannii isolated from burn wards. Burns. 2014;40(2):295-9. [PubMed ID: 23886986]. https://doi.org/10.1016/j.burns.2013.06.003.
-
37.
Peymani A, Higgins PG, Nahaei MR, Farajnia S, Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int J Antimicrob Agents. 2012;39(6):526-8. [PubMed ID: 22521767]. https://doi.org/10.1016/j.ijantimicag.2012.02.014.
-
38.
Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538-82. [PubMed ID: 18625687]. https://doi.org/10.1128/CMR.00058-07.
-
39.
Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, El Kholy A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int J Infect Dis. 2014;22:49-54. [PubMed ID: 24607428]. https://doi.org/10.1016/j.ijid.2013.12.004.
-
40.
Bahador A, Raoofian R, Farshadzadeh Z, Beitollahi L, Khaledi A, Rahimi S, et al. The Prevalence of IS Aba 1 and IS Aba 4 in Acinetobacter baumannii Species of Different International Clone Lineages Among Patients With Burning in Tehran, Iran. Jundishapur J Microbiol. 2015;8(7):17167. [PubMed ID: 26396712]. https://doi.org/10.5812/jjm.17167v2.