Abstract
Background:
Diarrhea continues to be one of the most common causes of morbidity and mortality among children in developing countries. Recently, some studies have implicated diffusely adherent Escherichia coli (DAEC) strains as a cause of diarrhea. The clinical manifestations of diarrhea caused by DAEC strains may include watery or bloody diarrhea, abdominal pain, dehydration, and fever.Objectives:
The aims of this study were (1) isolation of E. coli from fecal samples obtained from patients with diarrhea in Shiraz (Iran), (2) detection of DAEC pathotypes in isolates by molecular diagnostic techniques such as conventional PCR and real-time PCR assays, and (3) investigation of the antibiotic susceptibility of DAEC isolates.Materials and Methods:
Seven hundred and fifteen stool specimens of diarrhea patients were collected in Shiraz, Fars, Iran. Diarrheagenic E. coli strains were isolated by standard biochemical analysis. Susceptibility testing was performed by the diffusion method, according to the guidelines of the Clinical and Laboratory Standards Institute. Real-time PCR and conventional PCR were used to detect the daaD gene in the DAEC strains isolated.Results:
Of the 715 stool samples tested, 101 (14.1%) were identified as E. coli by biochemical tests and culture. Of the infected patients, 58 (57%) were male and 43 (43%) were female, with a mean age of 13.52 (SD, 1.66) years. Eight of the E. coli strains were identified as DAEC strains. The most effective antibiotics against the DAEC isolated were levofloxacin and imipenem, and the least effective antibiotics were ampicillin and penicillin.Conclusions:
Our analysis indicated that DAEC strains may be considered as potential pathogens in Shiraz, southern Iran. Further, although the prevalence of DAEC is low, prevention of infection caused by this bacterium among asymptomatic patients is crucial. Therefore, further characterization of the different virulence aspects of DAEC strains is required.Keywords
Diarrhea Diffusely Adherent E. coli (DAEC) Children Real-time PCR
1. Background
Pathogenic Escherichia coli strains are classified into pathotypes on the basis of their virulence genes. Based on the pathogenesis, diarrheagenic E. coli (DEC) are classified into enteroaggregative E. coli, enterotoxigenic E. coli, enterohemorrhagic E. coli, enteropathogenic E. coli and enteroinvasive E. coli (1). Blanco (2006) reported that diffusely adherent E. coli (DAEC) are also as a diarrheagenic group of E. coli (2). DAEC strains are defined by their diffuse adherence pattern on cultured HeLa or HEp-2a cells, in which the bacteria uniformly cover the entire cell surface. The presumed virulence factor of DAEC strains is diffuse attachment to the intestinal lining of the infected host. DAEC binding induces the recruitment of DAF and CEA family receptors around the adherent bacteria via a lipid raft-dependent mechanism, which initiates internalization and cell signaling. The Afa/Dr family of adhesions in DAEC strains may be responsible for the adherence phenotype (3). In addition, the prevalence of afaC-positive DAEC isolates is high in the colonic mucosa in colon cancer (4). In developing countries, DAEC strains may cause diseases in malnourished or immunologically naive children (5). However, the pathogenicity and epidemiologic significance of DAEC isolates has long been the subject of controversy.
Food or water contaminated with human or animal feces is the main transmission route for DAEC pathovars. Clinical manifestations of diarrhea episodes caused by diarrhea-causing DAEC may include watery or bloody diarrhea, abdominal pain, dehydration, and fever; however, there is no unique clinical characteristic that is specific to DAEC intestinal infections (6). Further, the prevalence of DAEC in the stool specimens of children with diarrhea is lower than that of other DEC pathotypes. Some studies have associated DAEC strains with diarrhea in infants, children and adults (7).
Some of the investigations on DAEC have shown that the bacterium may be present in children without clinical symptoms, while other studies have shown a relationship between DAEC infection and the presence of symptoms. DAEC-related acute diarrhea is a relatively new topic that is of public health significance (1, 8).
A fimbrial adhesin designated F1845 has been shown to be responsible for the diffuse cell adherence of a diarrheal E. coli isolate. Further, a polypeptide with an apparent size of 15.5 kDa has been shown to be encoded by the daaD gene, which is believed to be the F1845 determinant (9).
There are very few studies on the epidemiology and prevalence of DAEC infections in Shiraz, Iran. Therefore, there is a need for more research on the epidemiology of DAEC infections for better treatment planning and prevention of diarrhea.
2. Objectives
The goals of this study were: 1) isolation of E. coli from patients with diarrhea in Shiraz (Iran), 2) detection of DAEC pathotypes in isolates by molecular diagnostic techniques such as conventional PCR and real-time PCR assays, and 3) investigation of the antibiotic susceptibility of the DAEC isolates.
3. Materials and Methods
3.1. Clinical Specimens and Culture Method
This cross-sectional study was performed from January 2012 to December 2013 in Shiraz (Iran). In total, 715 stool samples from diarrhea patients were collected from different hospitals in Shiraz. The fecal samples were transported to the laboratory using the Cary Blair transport medium. Demographic data such as age, gender and clinical findings were obtained from the physicians. Fever, diarrhea and vomiting were the common clinical symptoms. The fecal swabs of the patients were streaked on MacConkey agar, after which the plates were incubated at 37°C for 24 hours. If pink colonies appeared, they were sub-cultured on eosin methylene blue so that they turned green in color. The isolates were identified as E. coli based on Gram staining, the oxidase test, catalase test, motility test, triple-sugar iron fermentation, the citrate test, methyl red staining, the Voges-Proskauer test, indole test, ortho-nitrophenyl-galactoside test, and acid production from carbohydrates (10, 11). For long-term storage, the purified isolates were stored in tryptic soy broth containing 20% glycerol (Merck Co., Germany) at -20°C.
3.2. Antibiotic Susceptibility Testing
Susceptibility testing for 18 antimicrobial agents (MAST Co., UK) was performed: imipenem (IMI), ciprofloxacin (CIP), levofloxacin (LEV), trimethoprim-sulfamethoxazole (SXT), amikacin (AK), gentamicin (GM), streptomycin (S), ceftriaxone (CRO), cefixime (CFM), cefotaxime (CTX), nalidixic acid (NA), tetracycline (T), azithromycin (AZT), ampicillin (AP), chloramphenicol (C), clarithromycin (CLA), penicillin (PG), and nitrofurantoin (NI). Susceptibility testing was performed using the diffusion method, according to the guidelines of the Clinical and Laboratory Standards Institute (12, 13). In this study, E. coli ATCC 25922 was used for quality control.
3.3. Molecular diagnostic methods
3.3.1. DNA extraction and primer selection
A sweep of three colonies were inoculated in Luria-Bertani broth containing 1% tryptone, 0.5% yeast extract, and 0.5% NaCl, and the broth was then incubated at 37°C overnight along with shaking. E. coli strains isolated from the samples were grown on Luria-Bertani agar (Sigma, St. Louis, MO) overnight at 37°C. E. coli DNA was extracted using the DNA extraction kit (QIAGEN Ltd., Crawley, UK), according to manufacturer’s instructions. The specific set of primers used to detect daaD in a single reaction is shown in Table 1 (14, 15).
Primers Used in the PCR Assays for the Detection of the daaD Gene
| Pathotype | Gene | Primer Sequence (5’ – 3’) | Amplicon Size (bp) (PCR) | Amplicon Tm (Real-Time PCR) |
|---|---|---|---|---|
| DAEC | daaD | F: TGAACGGGAGTATAAGGAAGATG | 371 | 93.2 |
| R: GTCCGCCATCACATCAAAA |
3.3.2. Real-Time PCR Assay
The real-time PCR assay was used to detect DAEC strains in a final reaction volume of 25 µL that contained 1 µL of SYBR Green I (Invitrogen, USA). The reactions were performed on Rotor-Gene 6000 (Corbett research, Australia) under the following conditions: 95°C for 5 minutes, followed by 45 cycles of 95°C for 30 seconds and 58°C for 40 seconds. Finally, melting curve analysis was performed from 80°C to 95°C with a ramping rate of 2.5°C/s, and fluorescence analysis was conducted every 2°C for 5 seconds. All the reactions were repeated in triplicate, and positive and negative control samples were used in each run. All the data were analyzed by the Rotor-Gene 6000 software, version 1.7 (Figure 1) (14, 15).
Identification of DAEC Strains by Real-Time PCR Assay of the daaD Virulence Gene

3.3.3. PCR Assay
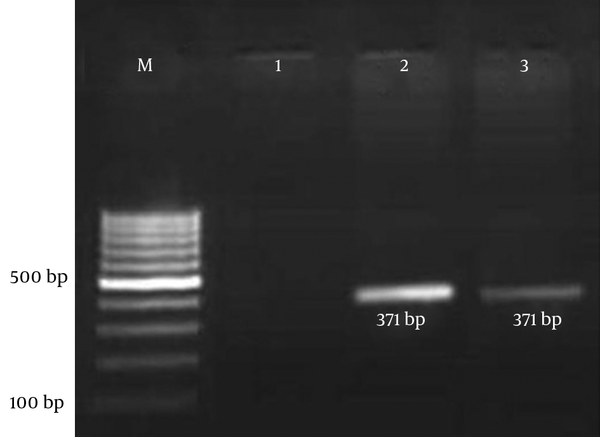
Each PCR assay was performed with a final reaction volume of 25 µL containing 2 µL of the template DNA, 200 mM deoxynucleoside triphosphates, 4 mM MgCl2, 1.5 U Taq DNA polymerase (CinnaGen, Iran), and 0.2 mM of each primer. The cycling parameters used were as follows: 95°C for 5 minutes for initial denaturation of the DNA; this was followed by 35 cycles of 1 minute at 94°C, 1 minute at 58°C, and 1 minute at 72°C; and a final single prolonged extension at 72°C for 5 minutes. A negative control was included in each experiment to eliminate the possibility of reagent contamination. The E. coli strain ECOR 50 was used as a positive control in the PCR test. The amplified product was visualized by gel electrophoresis in 1.5% agarose gel containing ethidium bromide for 45 minutes at 100 V and then visualized under UV light (Figure 2) (14, 15).
Agarose Gel Electrophoresis of Amplicons from the PCR
3.4. Data Analysis
The Ax2 test or Fisher’s exact test was used to verify the significance of the data. A P value < 0.05 was considered to indicate statistical significance.
4. Results
In the present study, 715 stool samples were examined, from which E. coli was identified in 101 (14.1%) samples by biochemical tests and culture. The sample population comprised 58 (57%) males and 43 (43%) females with a mean age of 13.52 (SD, 1.66) years (range, 2 months to 63 years). No significant correlation was observed between E. coli infection and gender (P = 0.141). DEC strains were isolated much more frequently in the summer months than in other seasons.
Molecular methods (real-time PCR and PCR) were applied to investigate the presence of the daaD gene in the 101 E. coli strains isolated from the diarrheal stool samples. Out of the 101 DEC strains identified, eight were confirmed as DAEC strains by real-time PCR. Seven DAEC strains were isolated from watery diarrhea samples, and 1 DAEC strain was isolated from a bloody diarrhea sample (Table 2). Three of the DAEC samples belonged to children aged between 6 and 12 months. Moreover, five DAEC strains were isolated from adults older than 50 years.
Distribution of DAEC Strains According to Season and Clinical Symptoms
| Clinical and Other Characterization | Number of DAEC Strains, No. (%) |
|---|---|
| Season | |
| Spring | 2 (25) |
| Summer | 4 (50) |
| Fall | 2 (25) |
| Winter | Not seen (0) |
| Clinical symptoms | |
| Watery diarrhea | 7 (87.5) |
| Bloody diarrhea | 1 (12.5) |
| Fever | 3 (37.5) |
| Vomiting | 6 (75) |
| Sex ratio (M/F) | |
| Male | 5 (62.5) |
| Female | 3 (37.5) |
The highest number of DAEC strains were isolated from watery diarrhea samples, but there was no significant correlation between the type of diarrhea and the presence of DAEC strains (P = 0.682) (Table 2). The most effective antibiotics against DAEC were levofloxacin and imipenem, while the least effective antibiotics were ampicillin and penicillin.
Susceptibility of DAEC Strains Isolated from the Stools of Patients with Diarrhea in Shiraz
| Antimicrobial agent | DAEC (n = 8) | ||
|---|---|---|---|
| Susceptible (%) | Intermediate (%) | Resistant (%) | |
| Cefotaxime | 62.5 | 12.5 | 25 |
| Ceftriaxone | 37.5 | 62.5 | 0 |
| Cefixime | 37.5 | 25 | 37.5 |
| Imipenem | 87.5 | 12.5 | 0 |
| Levofloxacin | 87.5 | 12.5 | 0 |
| Ciprofloxacin | 75 | 25 | 0 |
| Nalidixic Acid | 37.5 | 25 | 37.5 |
| Chloramphenicol | 50 | 25 | 25 |
| Tetracycline | 37.5 | 62.5 | 0 |
| Trimethoprim/Sulfamethoxazol | 37.5 | 25 | 37.5 |
| Streptomycin | 37.5 | 37.5 | 25 |
| Gentamicin | 62.5 | 37.5 | 0 |
| Amikacin | 62.5 | 25 | 12.5 |
| Nitrofurantoin | 62.5 | 25 | 12.5 |
| Ampicillin | 12.5 | 25 | 62.5 |
| Penicillin | 37.5 | 0 | 62.5 |
| Azithromycin | 75 | 0 | 25 |
| Clarithromycin | 75 | 12.5 | 12.5 |
5. Discussion
DAEC strains are a newly proposed virotype of diarrheagenic E. coli (15). Recently, some studies have implicated DAEC strains as a cause of diarrhea, whereas some studies have indicated that the identification of DAEC strains is more common in asymptomatic controls than in diarrhea patients (16-18). Further, one report in Brazil (2008) showed that DAEC strains were isolated in 18.3% of children with diarrhea (8).
In this study, DAEC strains were isolated in 8 (3.92%) patients with diarrhea. This is the first study performed in Shiraz to identify DAEC intestinal pathogens in diarrhea patients. The frequency and other epidemiological characteristics of this pathogen as the causative agent of diarrhea differ from district to district. This variation has also been reported in countries in similar geographical regions (19, 20). Recently, several of these potential virulence characteristics have been identified. Additionally, in a few other studies, DAEC strains have been identified as the cause of diarrhea only in patients older than six months and in adults (21).
According to a study in Brazil, DAEC strains were detected in all age groups, but the frequency of detection was significantly higher in children over 1 year old (8). In other studies in France and New Caledonia, DAEC strains were significantly associated with diarrhea only in children over 24 months of age (3, 22). In yet another study, it was shown that the relative risk of diarrhea caused by DAEC increased with age, from 12 months of age to 5 years (17). DAEC has been implicated as the cause of diarrhea in several other studies, particularly in children >1 year old (3, 23, 24). In the present study, DAEC strains were isolated in patients from all age groups with diarrhea. Our findings are similar to those of studies from Caledonia, France and some other regions (4, 22-24).
In a study by Guion et al. (15), BLAST data indicated that the daaD gene was better conserved than the other closely linked genes related to the Dr adhesion phenotype (10). Further, daaD-positive DAEC was recently associated with disease in adults in Ghana in a study that employed a definitive methodology (25, 26). Thus, based on the findings of these previous studies, we chose the daaD gene to identify the strains of DAEC. PCR assay is a practical and rapid method for the identification of virulence genes and is commonly used for identifying E. coli strains (25), so this was our method of choice.
The limitations of biochemical and serological diagnostic methods can be overcome by molecular diagnostic assays, which are sensitive and specific (19). Real-time PCR, which is one such method, offers faster and more robust identification, and gel electrophoresis is not required to identify the amplification products after PCR (24, 27) The advantages of real-time PCR over traditional PCR include its closed tube system, which does not require post-PCR processing. Real-time PCR also offers higher precision, better sensitivity (down to 1 copy), a better dynamic range (greater than 8 logs) and higher resolution (less than two-fold difference) (28).
In the present study, a high percentage of the DAEC strains were resistant to ampicillin (62.5%), penicillin (62.5%), nalidixic acid (37.5%), cefixime (37.5%) and trimethoprim/sulfamethoxazol (37.5%). Alikhani also reported a higher percentage of resistance to ampicillin, cefotaxime and trimethoprim/sulfamethoxazol in E. coli (29). Another report from northern Iran (2014) showed that 79.4% of the isolated E. coli were resistant to cefixime (30). Studies from Peru, Vietnam, Brazil and Mexico have also reported greater resistance to ampicillin in E. coli (31-34). Our findings are similar to those of the studies from Iran, Peru, Vietnam, Brazil and Mexico (29-34). It should be also noted that in many cases, antibiotic resistance is transmitted to humans, hospitalized patients and the hospital environment through other sources, including food plants, animals, fish, poultries, and other industries, in which antibiotics are used for different purposes and may lead to the emergence of resistant strains (35-38). The isolation, identification and antimicrobial susceptibility of pathogens can be helpful in optimizing the use of antimicrobial agents.
In conclusion, our analysis indicated that DAEC strains may be considered as potential pathogens in Shiraz, southern Iran. The results showed that although the prevalence of DAEC is low, prevention of infections caused by this bacterium among asymptomatic patients is crucial. Therefore, further characterization of the different virulence aspects of DAEC strains is required.
Acknowledgements
References
-
1.
Meraz IM, Arikawa K, Nakamura H, Ogasawara J, Hase A, Nishikawa Y. Association of IL-8-inducing strains of diffusely adherent Escherichia coli with sporadic diarrheal patients with less than 5 years of age. Braz J Infect Dis. 2007;11(1):44-9. [PubMed ID: 17625726].
-
2.
Blanco M, Blanco JE, Dahbi G, Alonso MP, Mora A, Coira MA, et al. Identification of two new intimin types in atypical enteropathogenic Escherichia coli. Int Microbiol. 2006;9(2):103-10. [PubMed ID: 16835840].
-
3.
Scaletsky IC, Fabbricotti SH, Carvalho RL, Nunes CR, Maranhao HS, Morais MB, et al. Diffusely adherent Escherichia coli as a cause of acute diarrhea in young children in Northeast Brazil: a case-control study. J Clin Microbiol. 2002;40(2):645-8. [PubMed ID: 11825986].
-
4.
Ene PC, Abbey SD, Madubuattah CG, Okeke CU. Phenotypic assay of adherent E. coli strains using hep-2 cells on diarrheic children in Rivers State of Nigeria. Int J Biol Chem Sci. 2012;6(1):421-6.
-
5.
Scaletsky IC, Pedroso MZ, Oliva CA, Carvalho RL, Morais MB, Fagundes-Neto U. A localized adherence-like pattern as a second pattern of adherence of classic enteropathogenic Escherichia coli to HEp-2 cells that is associated with infantile diarrhea. Infect Immun. 1999;67(7):3410-5. [PubMed ID: 10377120].
-
6.
Gunzburg ST, Chang BJ, Elliott SJ, Burke V, Gracey M. Diffuse and enteroaggregative patterns of adherence of enteric Escherichia coli isolated from aboriginal children from the Kimberley region of Western Australia. J Infect Dis. 1993;167(3):755-8. [PubMed ID: 8440943].
-
7.
Daigle F, Harel J, Fairbrother JM, Lebel P. Expression and detection of pap-, sfa-, and afa-encoded fimbrial adhesin systems among uropathogenic Escherichia coli. Can J Microbiol. 1994;40(4):286-91. [PubMed ID: 7913657].
-
8.
Spano LC, Sadovsky AD, Segui PN, Saick KW, Kitagawa SM, Pereira FE, et al. Age-specific prevalence of diffusely adherent Escherichia coli in Brazilian children with acute diarrhoea. J Med Microbiol. 2008;57(Pt 3):359-63. [PubMed ID: 18287300]. https://doi.org/10.1099/jmm.0.47660-0.
-
9.
Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev. 2005;18(2):264-92. [PubMed ID: 15831825]. https://doi.org/10.1128/CMR.18.2.264-292.2005.
-
10.
Soltani J, Poorabbas B, Miri N, Mardaneh J. Health care associated infections, antibiotic resistance and clinical outcome: A surveillance study from Sanandaj, Iran. World J Clin Cases. 2016;4(3):63-70. [PubMed ID: 26989670]. https://doi.org/10.12998/wjcc.v4.i3.63.
-
11.
Poorabbas B, Mardaneh J, Rezaei Z, Kalani M, Pouladfar G, Alami MH, et al. Nosocomial Infections: Multicenter surveillance of antimicrobial resistance profile of Staphylococcus aureus and Gram negative rods isolated from blood and other sterile body fluids in Iran. Iran J Microbiol. 2015;7(3):127-35. [PubMed ID: 26668699].
-
12.
Clinical and Laboratory Standards Institute. Clinical and Laboratory Standards Institute (CLSI). M100-S21. 31. 2011.
-
13.
Mardaneh J, Dallal MM. Isolation, identification and antimicrobial susceptibility of Pantoea (Enterobacter) agglomerans isolated from consumed powdered infant formula milk (PIF) in NICU ward: First report from Iran. Iran J Microbiol. 2013;5(3):263-7. [PubMed ID: 24475334].
-
14.
Frommel U, Bohm A, Nitschke J, Weinreich J, Gross J, Rodiger S, et al. Adhesion patterns of commensal and pathogenic Escherichia coli from humans and wild animals on human and porcine epithelial cell lines. Gut Pathog. 2013;5(1):31. [PubMed ID: 24188314]. https://doi.org/10.1186/1757-4749-5-31.
-
15.
Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J Clin Microbiol. 2008;46(5):1752-7. [PubMed ID: 18322059]. https://doi.org/10.1128/JCM.02341-07.
-
16.
Gomes TA, Vieira MA, Abe CM, Rodrigues D, Griffin PM, Ramos SR. Adherence patterns and adherence-related DNA sequences in Escherichia coli isolates from children with and without diarrhea in Sao Paulo city, Brazil. J Clin Microbiol. 1998;36(12):3609-13. [PubMed ID: 9817882].
-
17.
Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez J, et al. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol. 1993;138(10):849-69. [PubMed ID: 8237973].
-
18.
Taddei CR, Moreno AC, Fernandes Filho A, Montemor LP, Martinez MB. Prevalence of secreted autotransporter toxin gene among diffusely adhering Escherichia coli isolated from stools of children. FEMS Microbiol Lett. 2003;227(2):249-53. [PubMed ID: 14592716].
-
19.
Abbasi P, Kargar M, Doosti A, Mardaneh J, Dehyadegari MA, Ghorbani-Dalini S. Multiplex Real-Time PCR Assay for the Detection of LT, STIa and STIb Genes in Enterotoxigenic Escherichia coli. Int J Enteric Pathog. 2014;2(1). https://doi.org/10.17795/ijep16431.
-
20.
Hegde A, Ballal M, Shenoy S. Detection of diarrheagenic Escherichia coli by multiplex PCR. Indian J Med Microbiol. 2012;30(3):279-84. [PubMed ID: 22885192]. https://doi.org/10.4103/0255-0857.99485.
-
21.
Mansan-Almeida R, Pereira AL, Giugliano LG. Diffusely adherent Escherichia coli strains isolated from children and adults constitute two different populations. BMC Microbiol. 2013;13:22. [PubMed ID: 23374248]. https://doi.org/10.1186/1471-2180-13-22.
-
22.
Germani Y, Begaud E, Duval P, Le Bouguenec C. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in new Caledonia. J Infect Dis. 1996;174(5):1124-6. [PubMed ID: 8896522].
-
23.
Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142-201. [PubMed ID: 9457432].
-
24.
Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123-40. [PubMed ID: 15040260]. https://doi.org/10.1038/nrmicro818.
-
25.
Okeke IN. Diarrheagenic Escherichia coli in sub-Saharan Africa: status, uncertainties and necessities. J Infect Dev Ctries. 2009;3(11):817-42. [PubMed ID: 20061678].
-
26.
Opintan JA, Bishar RA, Newman MJ, Okeke IN. Carriage of diarrhoeagenic Escherichia coli by older children and adults in Accra, Ghana. Trans R Soc Trop Med Hyg. 2010;104(7):504-6. [PubMed ID: 20307897]. https://doi.org/10.1016/j.trstmh.2010.02.011.
-
27.
Souza TB, Lozer DM, Kitagawa SM, Spano LC, Silva NP, Scaletsky IC. Real-time multiplex PCR assay and melting curve analysis for identifying diarrheagenic Escherichia coli. J Clin Microbiol. 2013;51(3):1031-3. [PubMed ID: 23303494]. https://doi.org/10.1128/JCM.02478-12.
-
28.
Abbasi P, Kargar M, Doosti A, Mardaneh J, Dalini SG, Dehyadegari MA. Real time pcr for characterization of enteroinvasive e. Coli (eiec) in children with diarrhea in shiraz. Ann Colorectal Res. 2014;2(3).
-
29.
Alikhani MY, Hashemi SH, Aslani MM, Farajnia S. Prevalence and antibiotic resistance patterns of diarrheagenic Escherichia coli isolated from adolescents and adults in Hamedan, Western Iran. Iran J Microbiol. 2013;5(1):42-7. [PubMed ID: 23466523].
-
30.
Esmaeili Dooki MR, Rajabnia R, Barari Sawadkohi R, Mosaiebnia Gatabi Z, Poornasrollah M, Mirzapour M. Bacterial entropathogens and antimicrobial susceptibility in children with acute diarrhea in Babol, Iran. Caspian J Intern Med. 2014;5(1):30-4. [PubMed ID: 24490011].
-
31.
Ochoa TJ, Ruiz J, Molina M, Del Valle LJ, Vargas M, Gil AI, et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am J Trop Med Hyg. 2009;81(2):296-301. [PubMed ID: 19635887].
-
32.
Nguyen TV, Le PV, Le CH, Weintraub A. Antibiotic resistance in diarrheagenic Escherichia coli and Shigella strains isolated from children in Hanoi, Vietnam. Antimicrob Agents Chemother. 2005;49(2):816-9. [PubMed ID: 15673777]. https://doi.org/10.1128/AAC.49.2.816-819.2005.
-
33.
Garcia PG, Silva VL, Diniz CG. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecal microbiota from children with and without acute diarrhea. J Microbiol. 2011;49(1):46-52. [PubMed ID: 21369978]. https://doi.org/10.1007/s12275-011-0172-8.
-
34.
Estrada-Garcia T, Cerna JF, Paheco-Gil L, Velazquez RF, Ochoa TJ, Torres J, et al. Drug-resistant diarrheogenic Escherichia coli, Mexico. Emerg Infect Dis. 2005;11(8):1306-8. [PubMed ID: 16102327]. https://doi.org/10.3201/eid1108.050192.
-
35.
Mardaneh J, Soltan-Dallal MM. Isolation and Identification of E. cowanii from Powdered Infant Formula in NICU and Determination of Antimicrobial Susceptibility of Isolates. Iran J Pediatr. 2014;24(3):261-6. [PubMed ID: 25562018].
-
36.
Anvarinejad M, Pouladfar GR, Pourabbas B, Amin Shahidi M, Rafaatpour N, Dehyadegari MA, et al. Detection of Salmonella spp. with the BACTEC 9240 Automated Blood Culture System in 2008 - 2014 in Southern Iran (Shiraz): Biogrouping, MIC, and Antimicrobial Susceptibility Profiles of Isolates. Jundishapur J Microbiol. 2016;9(4):26505. [PubMed ID: 27284396]. https://doi.org/10.5812/jjm.26505.
-
37.
Mardaneh J, Soltan Dallal MM, Taheripoor M, Rajabi Z. Isolation, Identification and Antimicrobial Susceptibility Pattern of Tatumella ptyseos Strains Isolated From Powdered Infant Formula Milk Consumed in Neonatal Intensive Care Unit: First Report From Iran. Jundishapur J Microbiol. 2014;7(6):10608. [PubMed ID: 25371802]. https://doi.org/10.5812/jjm.10608.
-
38.
Shaghaghian S, Pourabbas B, Alborzi A, Askarian M, Mardaneh J. Vancomycin-Resistant Entrococci colonization in chronic hemodialysis patients and its risk factors in southern Iran (2005-2006). Iran Red Crescent Med J. 2012;14(10):686-91. [PubMed ID: 23285424].