Abstract
Background:
Methicillin-resistant Staphylococcus aureus (MRSA) is considered one of the most important pathogenic bacteria and most prevalent pathogens causing dangerous infections in humans.Objectives:
The purpose of this study was to analyze the hypervariable region (HVR) diversity of clinical MRSA isolates in Tabriz, northwestern Iran.Methods:
In this retrospective and descriptive study, from Staphylococcus aureus strains isolated from clinical specimens of hospitalized patients from 2006 to 2013 at Tabriz health centers, 151 isolates were randomly selected. Methicillin-resistant isolates were identified by the agar disk diffusion method and mecA PCR assays. The genetic diversity of the isolates in the HVR were analyzed with the HVR typing method.Results:
According to the antibiogram test results, from 151 samples, 52 isolates (34.4%) were resistant to cefoxitin. However, based on the polymerase chain reaction (PCR) assay, 54 isolates (35.8%) had the mecA gene and were identified as MRSA strains. According to PCR of the mecHVR, these MRSA strains were classified into seven different genotypes of HVR groups.Conclusions:
High HVR diversity among the studied MRSA isolates could be a result of insufficient or inadequate infection-control protocols in Tabriz hospitals. Moreover, the high number of HVR genotypes showed that HVR typing can be used along with other typing methods in epidemiological studies of MRSA as a useful tool for monitoring, tracking contaminations, and controlling infections in hospital settings.Keywords
1. Background
Staphylococcus aureus is considered one of the most important pathogenic bacteria and most prevalent pathogens that cause dangerous infections in humans. Approximately one third of healthy people are colonized by S. aureus without any illness. However, S. aureus can cause various diseases, ranging from minor superficial infections to acute and intense infections, such as osteomyelitis, bacteremia, and endocarditis (1, 2). Moreover, the development and spread of antibiotic resistance among clinical isolates of S. aureus, especially the emergence of methicillin-resistant S. aureus (MRSA) strains, which are resistant to many different groups of antimicrobial agents, is a growing problem that can lead to increased and longer hospital stays, more complicated treatment, greater mortality, and higher healthcare costs (3). MRSA was first reported in England in 1961 (4), and its prevalence increased rapidly throughout different parts of the world. These strains, in addition to being resistant against beta-lactams, sometimes cause multidrug-resistant infections, especially in hospitalized patients (5-7). Therefore, continuous local monitoring of their prevalence and identifying different clones of these isolates in communities, especially in medical centers, is of great importance. To investigate the epidemiology of pathogenic bacteria, choosing an effective, simple, and accurate method is very important for identifying the bacterium’s origin and tracing its development and spread (7).
Different phenotypic and molecular methods, such as antibiotyping, phage typing, and pulsed-field gel electrophoresis, have been used in recent decades to identify, type, and track infections caused by S. aureus. In recent years, developments in molecular biology have led to the replacement of previous phenotypic methods with genotyping methods that have higher sensitivity and specificity, including multilocus sequence typing (MLST), multilocus variable-number tandem-repeat (VNTR) analyses, and other rapid methods that rely on polymerase chain reaction (PCR) (7-11).
One of the proposed typing methods involves hypervariable region (HVR) proliferation, or HVR typing, for MRSA strains through PCR in the investigation for polymorphisms in this region (12). The DNA sequence between IS431mec and the mecA gene, a region responsible for resistance against methicillin, is known as the HVR. The mec HVR is composed of direct repeated units, each with a size of almost 40 base pairs. However, since the number of these repeated units may be different between isolates, the HVR region can be used to type and classify MRSA strains. Compared to other molecular methods, HVR typing of MRSA offers greater speed and facility (12).
In recent years, several cases of MRSA isolates with reduced susceptibility to conventional antibiotics, such as vancomycin, have been reported in different parts of Iran, so the consistent monitoring of the diversity of isolated strains in each region is necessary in order to develop appropriate strategies for identifying their origin and preventing their spread. To the best of our knowledge, no investigation has previously been carried out on MRSA strain typing in northwestern Iran.
2. Objectives
The present investigation was designed and carried out in Tabriz hospitals to determine the diversity of clinical MRSA isolates based on the mecHVR.
3. Methods
In this retrospective and descriptive study carried out at Tabriz University of Medical Sciences, 151 non-repetitive clinical isolates of S. aureus were randomly selected. The included samples were selected from stock samples isolated over eight years (December 2006 to December 2013) from hospitalized patients in Tabriz medical centers. They were stored at -70°C in trypticase soy broth (Liofilchem, Italy) containing 15% glycerol (13). After the samples were thawed at laboratory temperature, they were incubated for 24 h at 37°C. The S. aureus isolates were identified based on colony morphology, gram staining, catalase, clamping factor, coagulase, growth in mannitol salt agar, and DNase tests (14).
Detection of methicillin resistance in S. aureus isolates was carried out with the disc agar diffusion test using a 30 μg cefoxitin disc (Code SD041, Hi-Media, Mumbai, India) on Mueller-Hinton agar (Hi-Media, Mumbai, India) containing 2% salt (NaCl), and the results were interpreted according to CLSI standards (15). S. aureus ATCC 29213 was used as a standard strain for quality-control of the susceptibility testing.
Chromosomal DNA extraction of the isolates was carried out with the CTAB method (16). For detection of the mecA gene among the studied isolates and evaluation of HVR polymorphisms in the MRSA strains, PCR was used with specific primers (Bioneer Co., Korea). All target genes and corresponding primers used for PCR amplification are listed in Table 1.
Primers Used for Amplification of Genes
| Gene | Primers Sequence (Forward and Reverse) (5’→3’) | Amplicon Size (bp) | Annealing Temp (°C) |
|---|---|---|---|
| mecA | 533 | 55 | |
| F | AAA ATC GAT GGT AAA GGT TGG C | ||
| R | ATG TCT GCA GTA CCG GAT TTG C | ||
| hvr | Variable | 55 | |
| F | ACT ATT CCC TCA GGC GTC C | ||
| R | GGA GTT AAT CTA CGT CTC ATC |
The PCR assays were accomplished in a 25 μL reaction mixture (consisting of Taq DNA polymerase, PCR buffer, MgCl2, dNTPs, primers, and template DNA) with an automated thermal cycler (Eppendorf Mastercycler gradient, Germany). All PCR components were provided by Cinnagen Co., Tehran, Iran. The PCR cycling conditions were as follows: initial denaturation at 94°C for 5 minutes, followed by 35 cycles at 94 °C for 1 minute, 55°C for 30 s, 72°C for 1 minute, and a final extension at 72°C for 3 minutes and 30 s. PCR products were separated by electrophoresis in 1.5% agarose gels (Sigma, CAS No. 9012-36-6) (12, 17).
In this study, the S. aureus ATCC 33591 and S. aureus ATCC 25923 strains were used as positive and negative controls, respectively, for the mecA gene. The data obtained were analyzed using SPSS version 16. We used descriptive statistical methods, including frequency and percentage.
4. Results
In this investigation, we used 151 random and non-duplicate isolates of S. aureus collected from clinical specimens of hospitalized patients in Tabriz hospitals over an 8-year period. Among the studied isolates, 62 (41%), 52 (34.4%), 16 (10.6%), 6 (4%), 6 (4%), 5 (3.3%), 2 (1.3%), and 2 (1.3%) were obtained from wounds, blood, urine, synovial fluid, sputum, endotracheal secretions, bile aspirates, and venous catheters cultures, respectively.
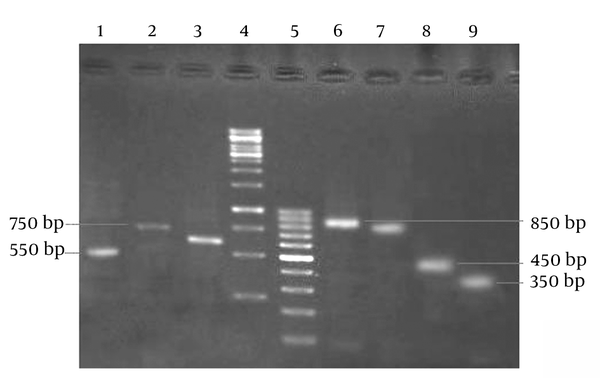
Of the S. aureus isolates, 52 (34.4%) were identified as MRSA based on resistance against the cefoxitin disc, while the other 99 isolates (65.6%) were sensitive against this antibiotic and thus classified as methicillin-sensitive S. aureus (MSSA) strains. Based on our results, the highest rates of MRSA isolates were collected from urine (9 of 16 isolates), catheters (1 of 2 isolates), endotracheal secretions (2 of 5 isolates), blood (19 of 52 isolates), sputum (2 of 6 samples), wounds (18 of 62 isolates), and synovial fluid (1 of 6 isolates), respectively. Based on PCR, the mecA gene was detected in 54 (35.8%) isolates. Indeed, only two isolates had no agreement with the disc diffusion results. All 54 mentioned isolates were used as MRSA strains for PCR in the HVR polymorphism investigation. HVR typing showed seven different patterns of the mec HVR among our MRSA isolates (Table 2). Figure 1 shows the electrophoresis results of different HVR patterns obtained from PCR products.
Characteristics of Different Patterns Obtained From HVR Proliferation in MRSA Isolates
| HVR Pattern | Amplicon Size (bp) | Number of Isolates (%) |
|---|---|---|
| H1 | 550 | 9 (16.7) |
| H2 | 750 | 15 (27.8) |
| H3 | 650 | 6 (11.1) |
| H4 | 850 | 5 (9.2) |
| H5 | 800 | 13 (24.1) |
| H6 | 450 | 3 (5.5) |
| H7 | 350 | 3 (5.5) |
Electrophoresis of HVR PCR Products on 1.5% Agarose Gel
5. Discussion
S. aureus, especially MRSA, is one of the most challenging human pathogens worldwide. Over the last few decades, this bacterium has been one of the most important causative agents for infections acquired in hospitals and the social environment. Thus, continuous monitoring of MRSA strains is necessary to understand the clonal evolution of successful lineages, which can guide the correct preventive decision-making in the control of infections. Therefore, the aim of this investigation was to determine the genotypic patterns of MRSA strains isolated from clinical specimens in Tabriz hospitals.
In this investigation, we used two different methods (evaluation of resistance against cefoxitin and identification of the mecA gene) in order to separate MRSA and MSSA strains. Comparing these two methods, the sensitivity of the phenotypic method with cefoxitin discs is relatively lower than with the genotypic method of mecA gene identification. However, even with the former method, the frequency of MRSA strains in this study was determined to be 35.8%, which is comparable to the rate found by other investigators in Iran (18, 19). Nevertheless, this amount, compared to the results of previous studies, with a prevalence 80% in Tabriz (20), is different. In another study carried out in Tehran by Maleki et al. (2006) (21), using 100 clinical isolates of S. aureus, the resistance rate against methicillin was reported to be 42%. The same year, Aligholi et al. (1) reported resistance in 47% of 338 S. aureus isolates collected from Tehran. In another study carried out in 2006 by Rahimi et al. (22) on 321 S. aureus isolates in Tehran, the MRSA prevalence was 88%. It seems that the main reason for the difference observed between the results of the present study and previous studies is that the time period was longer in this study. Only 1-year isolates were used in the other studies, but the present investigation used a collection of isolates from an 8-year period.
On the other hand, different MRSA prevalence rates have been published in different parts of the world. For example, Oguri et al. (23) reported that the MRSA prevalence was 61.9% in Japan in 2000. Li et al. (24) reported that the MRSA prevalence in China in 2007 was 82.5% (25), and a study carried out in Argentina showed a prevalence of 52%.
Unlike two previous studies (20, 26), it was observed in this study that there is not a significant correlation between MRSA strains and staphylococcal septicemia. Moreover, based on our results, seven different patterns of the HVR region were identified among MRSA strains. This finding is in complete agreement with Salmenlinna et al.’s (27) observations in 2001 using 72 MRSA strains collected from Finland, which identified seven HVR types. However, Senna et al. (28) carried out a study in Brazil in 2001 in which PFGE and HVR typing methods were compared for the typing of 97 MRSA isolates; based on the results, four HVR types were identified, which was less diverse compared to our study. In another investigation, Schmitz et al. (29) typed 183 S. aureus isolates by using the HVR method and other typing methods, and identified five HVR patterns among the MRSA strains, less diverse compared to our study. In 2005, Corrente et al. (30) compared two methods, HVR and RAPD, for the typing of MRSA strains in Italy and found five HVR patterns in 71 isolates. In Iran, Bagherzadeh Yazdchi et al. (12) compared antibiotyping and HVR typing methods in 64 strains of MRSA isolated from Tehran hospitals, and ten HVR patterns were found.
Considering the abovementioned issues, it can be concluded that compared to other countries, which might have better conditions for observing sanitation and health principles, MRSA strain typing in Iran will indicate more diversity. This can be considered criteria for demonstrating a more diverse clonal spread and placement of MRSA in hospitals.
Finally, based on the results obtained in the present study, HVR typing can be used along with other molecular methods as an appropriate technique in epidemiological investigations to control and monitor infections obtained at hospitals and from social contact, due to its higher strength in distinguishing MRSA isolates collected from clinical specimens. Furthermore, the high HVR diversity among the MRSA isolates showed that the infection-control protocols used in Tabriz hospitals lacked the necessary competence.
Acknowledgements
References
-
1.
Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Abdolmaleki Z, Sedaghat H, et al. Antibiotic susceptibility pattern of Gram-positive cocci cultured from patients in three university hospitals in Tehran, Iran during 2001-2005. Act Med Iran. 2009;47(4):329-34.
-
2.
Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A. VanA and VanB Positive Vancomycin-resistant Staphylococcus aureus Among Clinical Isolates in Shiraz, South of Iran. Oman Med J. 2014;29(5):335-9. [PubMed ID: 25337309]. https://doi.org/10.5001/omj.2014.90.
-
3.
Rice LB. Antimicrobial resistance in gram-positive bacteria. Am J Infect Control. 2006;34(5 Suppl 1):11-9. [PubMed ID: 16813977]. https://doi.org/10.1016/j.ajic.2006.05.220.
-
4.
Navidinia M, Fallah F, Lajevardi B, Shirdoost M, Jamali J. Epidemiology of methicillin-resistant Staphylococcus aureus isolated from health care providers in Mofid children hospital. Arch Pediatr Infect Dis. 2015;3(2):16458.
-
5.
Livermore D. Antibiotic resistance in staphylococci. Int J Antimicrob Agents. 2000;16(1):3-10.
-
6.
Hirmatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997;40(7):135-6.
-
7.
Oliveira DC, Tomasz A, de Lencastre H. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect Dis. 2002;2:180-9.
-
8.
Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99(11):7687-92. [PubMed ID: 12032344]. https://doi.org/10.1073/pnas.122108599.
-
9.
Spratt BG. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Curr Opin Microbiol. 1999;2(3):312-6. [PubMed ID: 10383857]. https://doi.org/10.1016/S1369-5274(99)80054-X.
-
10.
Hallin M, Deplano A, Denis O, De Mendonca R, De Ryck R, Struelens MJ. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J Clin Microbiol. 2007;45(1):127-33. [PubMed ID: 17093021]. https://doi.org/10.1128/JCM.01866-06.
-
11.
Fateh Amirkhiz M, Ahangarzadeh Rezaee M, Hasani A, Aghazadeh M, Naghili B. SCCmec typing of methicillin-resistant Staphylococcus aureus: An eight year experience. Arch Ped Infect Dis. 2015;3(4):30632.
-
12.
Bagherzadeh Yazdchi S, Poormand M, Haji abdolbaghi M, Hoseini M, Mardani N. Molecular characterization of hypervariable region (hvr) and antibiotic susceptibility patterns of Staphylococcus aureus strains isolates collected from Tehran University of Medical Sciences Hospitals. J Sch Pub Healt Inst Pub Healt Res. 2008;26:39-47.
-
13.
Tille PM. Bailey & Scott's Diagnostic Microbiology. 13 ed. UK: Louis: W.B. Saunders Elsevier; 2014.
-
14.
Mahon CR, Lehman DC, Manuselis G. Textbook of Diagnostic Microbiology. 3 ed. UK: Louis: W.B. Saunders Elsevier; 2007.
-
15.
Performance standards for antimicrobial susceptibility testing; Twenty-Third informational supplement, CLSI document M100-S23. Pennsylvania,USA,: Clinical and Laboratory Standards Institute, Wayne; 2013.
-
16.
Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor, Cold Spring Harbor Laboratory Press. 2012.
-
17.
Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal ecrotizing pneumonia in young immunocompetent patients. Lancet. 2002;359:753-9.
-
18.
Poorabbas B, Mardaneh J, Rezaei Z, Kalani M, Pouladfar G, Alami MH, et al. Nosocomial Infections: Multicenter surveillance of antimicrobial resistance profile of Staphylococcus aureus and Gram negative rods isolated from blood and other sterile body fluids in Iran. Iran J Microbiol. 2015;7(3):127-35. [PubMed ID: 26668699].
-
19.
Rahimi F, Bouzari M, Katouli M, Pourshafie M. Prophage typing of methicillin resistant Staphylococcus aureus isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J Microbiol. 2012;6(1):80-5.
-
20.
Abdoli Oskouie S, Ahangarzadeh Rezaee M, Ajhangh A, Abdinia B. Antimicrobial resistance pattern and minimum inhibitory concentration of vancomycin among Staphylococcus aureus and coagulase-negative Staphylococci, isolated from clinical specimens of children in Tabriz. J Ardabil Univ Med Sci. 2013;13(1):24-34.
-
21.
Maleki Z,, Anjarani S. Comparision of disk diffusion and E-test methods for oxacillin and vancomycin. Azad Uni Med J. 2007;16(4):211-5.
-
22.
Rahimi F, Bouzari M, Maleki Z, Rahimi F. Antibiotic susceptibility pattern among Staphylococcus spp. with emphasis on detection of mecA gene in methicillin resistant Staphylococcus aureus isolates. Iran J Clin Infect Dis. 2009;4(3):143-50.
-
23.
Oguri T, Igari J, Hirmatsu K, Watanabe A, Inoue M, Abe M. Beta-lactamase-producing activity and antimicrobial susceptibility of major pathogenic bacteria isolated from clinical samples. Japan: Japan beta-lactamase Reserch Group; 2002.
-
24.
Li M, Zhang GA, Liu Y. [Analysis of predominant bacteria of burn infection and their resistance to antibiotics in recent years]. Zhonghua Shao Shang Za Zhi. 2007;23(2):91-3. [PubMed ID: 17649879].
-
25.
Sola C, Gribaudo G, Vindel A, Patrito L, Bocco JL, Cordoba MCSG. Identification of a novel methicillin-resistant Staphylococcus aureus epidemic clone in Cordoba, Argentina, involved in nosocomial infections. J Clin Microbiol. 2002;40(4):1427-35. [PubMed ID: 11923368].
-
26.
Hadadi A, Moradi-Tabriz H, Mehdipour Aghabagher B, Moslehi B, Esmaielzadeh P. Determining the prevalence of methicillin- and vancomycin-resistant Staphylococcus aureus by MIC and E-Test. Teh Uni Med J. 2011;69(6):344-51.
-
27.
Salmenlinna S, Vuopio-Varkila J. Recognition of two groups of methicillin-resistant Staphylococcus aureus strains based on epidemiology, antimicrobial susceptibility, hypervariable-region type, and ribotype in Finland. J Clin Microbiol. 2001;39(6):2243-7. [PubMed ID: 11376064]. https://doi.org/10.1128/JCM.39.6.2243-2247.2001.
-
28.
Senna JPM, Pinto CA, Carvalho LPS, Santos DS. Comparison of pulsed-field gel electrophoresis and PCR analysis of polymorphisms on the mec hypervariable region for tyoing methicillin-resistant Staphylococcus aureus. J Clin Micbiol. 2002;40(6):2254-6.
-
29.
Schmitz FJ, Steiert M, Tichy HV, Hofmann B, Verhoef J, Heinz HP, et al. Typing of methicillin-resistant Staphylococcus aureus isolates from Dusseldorf by six genotypic methods. J Med Microbiol. 1998;47(4):341-51. [PubMed ID: 9569001]. https://doi.org/10.1099/00222615-47-4-341.
-
30.
Corrente M, Monno R, Totaro M, Martella V, Buonavoglia D, Rizzo C, et al. Characterization of methicillin resistant Staphylococcus aureus (MRSA) isolated at the Policlinico Hospital of Bari (Italy). New Microbiol. 2005;28(1):57-65. [PubMed ID: 15782627].