Abstract
Keywords
1. Background
Human complexity is often identified by biologists, dentists, anthropologists, and chemists. Human identification using DNA profile is the most widely used technique in forensic medicine. The use of short tandem repeat (STR) sequences is an accurate method for identifying individuals (1). In many instances, tooth or bone samples are the only source of DNA for individual and kinship identification from degraded human remains (2, 3). Based on these sequences, commercial kits have been developed, which are used in forensic centers to identify humans. Natural disasters lead to the destruction or disintegration of corpses; the bacterial degradation of soft tissues (blood samples, muscle tissue) occurs in a relatively short period of time (4, 5). Any damage to DNA with STR sequences makes identifying the individual difficult. Since soft tissues and blood decay faster after death, lots of damages would be caused to the DNA structure. On the other hand, hard tissues such as bones and teeth result in the stability of cellular tissue against decay due to their structural nature. Teeth and their composition, as well as their location in the jaw, provide better conditions to protect DNA and are an important source of DNA extraction in most cases. Teeth and their chemical analysis, in addition to determining a person’s year of birth, can also determine the date of death in some cases (6). The use of hard tissues such as teeth is known as one of the most suitable sources of DNA for genetic profiling of individuals even after several years from death (7). However, not all tooth samples contain sufficient amounts or quality of genomic DNA for STR analysis. The contamination of forensic samples with exogenous human DNA because of mishandling during recovery or processing remains an issue in many analyses (8, 9).
DNA extraction is performed based on the release of the cell genome, the removal of protein and chemical contaminants from it, and the preparation of pure DNA (10). As we know, research on DNA extraction in forensic medicine is associated with problems because endogenous DNA is generally present in small amounts and different levels of degradation. Currently, there are many applications of DNA extraction methods from teeth that can recover DNA. Given this, several techniques have been published, all of which aim to maximize DNA yields (11, 12). In most of the conventional techniques for DNA extraction from teeth, the powdered tissue technique is used to extract DNA. These methods rely on grinding the samples into a fine powder to produce higher yields. However, the greater level of sample manipulation increases the risk for contamination. Selection of tooth depends on availability and many other logistical and environmental factors at the time of sampling.
Despite the increasing research conducted on tooth tissues in genetics, little reliable information has been published, and the results of various methods are limited Although many studies have been done in the field of bone tissue and extraction methods, few studies have been carried out in the field of teeth, and they do not have much depth. However, many methods have been proposed for tooth sampling. Some of these methods aim at sourcing pulp and/or dentine and include horizontal (13) or vertical sectioning (14) and regular (15) or apical (16) endodontic access, with subsequent scraping or drilling (17) of the interior of the tooth. Other methods simply grind up all or part of the tooth by crushing between two steel plates (17), or grinding with a mortar and pestle (13), bone mill, blender (18), tissue grinder (19), or cryogenic grinder (20).
Adler et al. (21) demonstrated that higher drill speeds had a negative impact on mtDNA recovery from ancient teeth and suggested that cutting or powdering should be performed at low speeds to reduce the heat and thus minimize the damage to the DNA.
Schwartz et al. (22) examined nuclear DNA yield from pulp recovered from teeth separated from the jaws. In this research, the teeth were subjected to changes in pH, temperature, moisture, and burial condition from periods one week to six months. Similarly, Alvarez Garcia et al. (23) also examined nuclear DNA yields from teeth separated from the jaws that were subjected to varying environments. Both experimental studies determined no significant effect of temperature on their ability to retrieve nuclear DNA. Perhaps the effects of temperature are more pronounced over longer time frames.
Tilotta et al. (24) compared the nuclear DNA from teeth crushed to those in which the pulp remnants were sampled via regular endodontic access and noted a significantly higher yield from the teeth sampled via the more conservative technique. In this instance, 90% and 70% success rates for mtDNA and nuclear DNA recovery from teeth, respectively, dating back to 300 BC and 1600 AD were reported.
A method allowing repatriation of teeth with the body after sampling was demonstrated by Shiroma (25). In this method, the tooth is cut horizontally by sampling the pulp and dentin through a perforated hole inside the tooth, which is connected with wax after the sampling is completed.
In hard tissues, due to the presence of calcium-rich structures, having access to the cell’s structure would be very difficult (19). One of the most common features of DNA extraction techniques is the removal of calcium from hard tissues. The use of different buffers and temperatures have been suggested for this purpose. Buffers containing ethylenediaminetetraacetic acid (EDTA) play an important role in removing or reducing the amount of calcium by removing dual-capacity ions like calcium. By removing calcium from hard tissues, DNA-containing cell’s structures would be available, and DNA can be extracted using conventional methods (26, 27).
2. Objectives
This study aimed to prepare DNA profiles from the dental samples of the volunteers and to compare the results with blood sample profiles. In this study, the method used to extract DNA from bone was also used for tooth, and a commercial kit (DNP kit Sina Clone co. Iran) was used to extract DNA from tissue. Thereafter, an EDTA-containing buffer and different temperatures were used to evaluate this method. To ensure proper use of this method, distilled water was also used for comparison.
3. Methods
3.1. Blood and Tooth Sample Collection
The tooth is a hard tissue that is highly resistant to environmental changes such as decay and temperature changes, and its DNA can be extracted for several minutes at high temperatures. However, due to corruption or burning of the body, the extraction of DNA from soft tissues leads to failure. Hard enamel and dentin coating protect the tooth pulp from the ingress of air currents and contamination. These reasons facilitate the extraction of DNA from the teeth. Hence, in this study, teeth were used to extract blood.
In this research, the wisdom teeth of 10 patients undergoing a surgery were collected in Falcon with 5 ml of sterile normal saline by obtaining personal consent. About 500 µL of blood was collected from the extracted tooth using a syringe by a dental surgeon and then stored in tubes containing EDTA anticoagulants.
3.2. DNA Extraction
One of the most important issues in performing polymerase chain reaction (PCR) and molecular studies is the correct purification of DNA from samples. To avoid errors and problems in the PCR process, care must be taken when extracting DNA from the sample to obtain DNA with the least contamination with protein or RNA. Extraction of at least one DNA sequence, which includes the region to be amplified, is one of the main conditions for a good extraction.
There are several enzymes in the cell that can break down DNA. By adding EDTA to the DNA extraction solution, the destructive action of these enzymes can be inhibited by chelating these ions. Therefore, the most suitable medium in DNA solution is the solution containing EDTA, which has the right amount of solutes, because DNA is more stable and soluble in saline solutions. The pH of the medium should be alkaline and mild (about 8), because the positive pH of histones decreases in this pH. Naturally, DNA in cells is a complex of DNA and proteins. In addition, cell proteins and lipids must be isolated from nucleic acid, in which case different compounds are used.
DNAs were extracted from blood samples according to the protocol of the DNP kit (Sina Clone co. Iran), and stored until use. The most important point in the extraction process is to avoid contamination of solutions and external devices that can be transmitted to the samples through the skin, sneezing, and coughing. Therefore, working with a mask and gloves is mandatory. Each tooth was thoroughly rinsed with a brush and sterile distilled water. Afterwards, the teeth were kept for 24 hours in the freezer at -70 ºC. Each tooth was separately powdered manually under a sterile condition under the hood using a mortar. In triplicate, 50 mg of tooth powder was incubated with 2 ml of EDTA buffer (0.5 mM) for 24 hours at 4°C, 18°C, 37°C, and 56°C in a shaker incubator. To evaluate the performance of the powder buffer, the tooth sample was incubated in triplicate for 24 hours at 56°C with 2 ml of sterile distilled water. The tube containing the buffer was then centrifuged at 10000 g for 3 minutes. Subsequently, the supernatant was discarded, and 100 µL of protease buffer was added to the precipitate, and the solution was dissolved with vertex. Next, 5 µL of protease K (20 mg/mL) was added to this solution, incubated for 3 hours at 55°C, and vortexed every 30 minutes. Then, the DNA samples were extracted according to the instructions of the kit (Sina Clone DNP).
Quantitative and qualitative characteristics of all the DNA samples obtained from blood and teeth were evaluated using DS-11 Series Spectrophotometer/Fluorometer device (Denovix USA), and the results were recorded. Then, 5 µL of all the samples were electrophoresed with 1% agarose gel to ensure the quality of the extracted DNA. After ensuring the quality of the extracted DNA samples, all the samples were used for genetic profiling following the instructions of the AmpFLSTR Identifier PCR Amplification kit (Applied Biosystems). Next, 1 µL of PCR product was mixed with 8.5 µL HiDi formamide and 0.5 µL internal size standard (Liz600), and injected into ABI 3500 genetic analyzer according to the laboratory protocol. The results of the analysis were evaluated with Genepamer IDX software and stored as the genetic profile file. All the results were matched with MOBIN GIS software, designed and implemented by a group of researchers of this center.
4. Results
The extracted DNA was evaluated at different temperatures using EDTA buffer and 50 mg of tooth powder. The average concentration of DNA extracted from the sample into ternary sort at 55°C was 19.68 ng/µL, and the standard deviation (SD) was 2.68. SD is a statistic that measures the dispersion of a dataset relative to its mean. A low standard deviation indicates that the values tend to be close to the mean of the set, while a high standard deviation indicates that the values are spread out over a wider range (28). At a 37°C, the concentration of DNA was equal to 12.23 ng/µL (SD = 1.76); at 22°C, it was equal to 17.19 ng/µL (SD = 2.59); and at 4°C, it was equal to 15.06 ng/µL (SD = 1.67). For evaluation, sterile distilled water was used instead of buffer, which was equal to 7.9 ng/µL (SD = 0.72) at 55°C. However, in gel electrophoresis, the extracted DNA did not have a specific band.
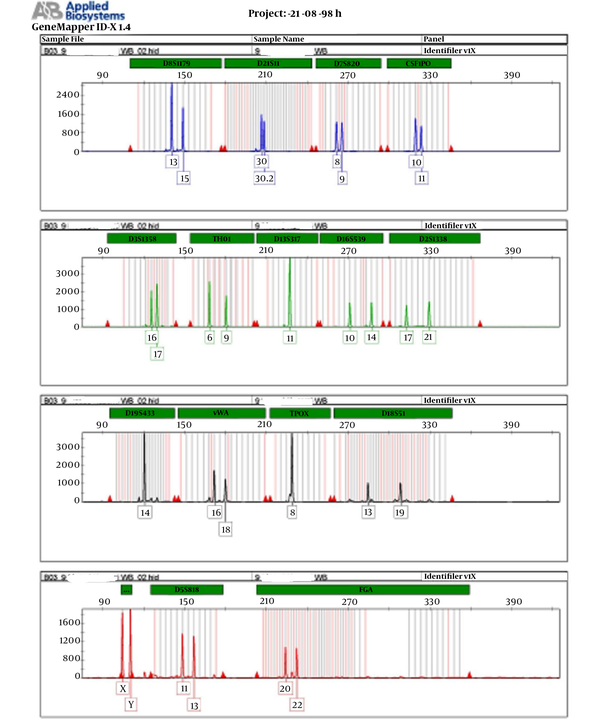
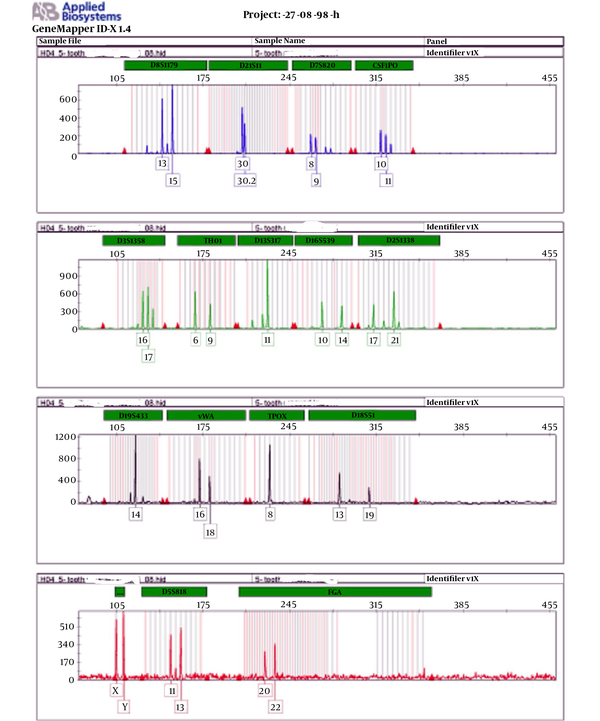
According to the instructions for preparing the genetic profile of Identifier, PCR was performed with 1 ng of extracted DNA. Profile results were evaluated based on different STR markers. To compare the results of two samples, each marker was evaluated for the presence of the same allele. The presence of one allele was considered as homozygous and two alleles as heterozygosis. The comparison of genetic profile alleles from blood and tooth samples showed complete agreement.
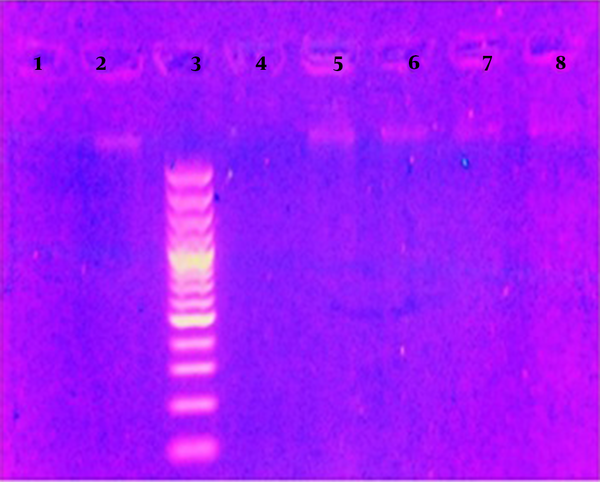
The results of this study are presented in Table 1, and the results of DNA quality extracted by electrophoresis are depicted in Figure 1. The results of the genetic profile of blood samples are presented in Figure 2, which shows each marker has one peak in the homozygous state and two peaks in the heterozygous state. The results of the genetic profile of teeth samples are presented in Figure 3, in which each marker has one peak in the homozygous state and two peaks in the heterozygous state.
| Buffer | Temperature, °C | DNA concentration, ng/µL | SD |
|---|---|---|---|
| EDTA buffer (0.5 mM) | 55 | 19.68 | 2.68 |
| EDTA buffer (0.5 mM) | 37 | 12.23 | 1.76 |
| EDTA buffer (0.5 mM) | 22 | 17.19 | 2.59 |
| EDTA buffer (0.5 mM) | 4 | 15.06 | 1.67 |
| dH2O | 55 | 7.9 | 0.72 |
The Results of DNA quality extracted by electrophoresis
The results of genetic profile of blood samples
The results of genetic profile of teeth samples
5. Discussion
The properties of teeth cause their stability and durability for many years. Hence, the use of teeth for archaeological and historical studies has been much considered by researchers in this field. Given these characteristics, the use of dental specimens for forensic studies as well as the identification of people involved in accidents and disasters whose tissues are no longer available would be helpful.
Due to the special texture of the teeth, the introduced method can be used. According to the results of this study, the use of EDTA buffer (0.5 mM) plays an important role in preparing samples for extraction. Notably, the temperature of this preparation is not significantly different from each other, and all cases can be used for the identification. In comparison with this buffer, although based on the results of determining the concentration with the nanodrop device the samples prepared by distilled water showed some errors, there was no band in electrophoresis. This indicated the ineffectiveness of this preparation method with distilled water. In various studies, different buffers have been used, of which EDTA has been considered as a component. Based on the results of the present study, it can be concluded that EDTA alone in the buffer is sufficient to prepare the right amount of DNA.
According to a previous study (27), this method was suitable for extraction from old bone samples. Although the temperature used in the mentioned method was 4°C, several temperatures were used in the present study, showing that at each temperature the appropriate amount of pure DNA can be obtained.
Since we used newly operated wisdom teeth, a complete concordance of the results of blood and tooth identification profiles in all 16 genetic markers was witnessed. However, for the teeth from a longer period, the number of evaluable genetic markers is usually limited and mini-filler kits could be used. This type of kit is suitable for evaluating genetic markers with short sequence length, with the main identification kit having 16 markers, and it is similar in only 9 markers.
5.1. Conclusions
This study showed that the buffer containing EDTA can be effective in the process of removing mineral elements in hard tissues such as bones and teeth and can provide the conditions for DNA extraction. Based on the results of this study, DNA can be extracted from the hard tissue through using a simple buffer containing EDTA. Since different temperatures had acceptable results, the DNA was extracted from hard tissues according to the facilities available in each laboratory.
References
-
1.
Dashnow H, Lek M, Phipson B, Halman A, Sadedin S, Lonsdale A, et al. STRetch: detecting and discovering pathogenic short tandem repeat expansions. Genome Biol. 2018;19(1):121. [PubMed ID: 30129428]. [PubMed Central ID: PMC6102892]. https://doi.org/10.1186/s13059-018-1505-2.
-
2.
Clayton TM, Whitaker JP, Maguire CN. Identification of bodies from the scene of a mass disaster using DNA amplification of short tandem repeat (STR) loci. Forensic Sci Int. 1995;76(1):7-15. [PubMed ID: 8591839]. https://doi.org/10.1016/0379-0738(95)01787-9.
-
3.
Alonso A, Martin P, Albarran C, Garcia P, Fernandez de Simon L, Jesus Iturralde M, et al. Challenges of DNA profiling in mass disaster investigations. Croat Med J. 2005;46(4):540-8. [PubMed ID: 16100756].
-
4.
Hochmeister MN, Budowle B, Borer UV, Eggmann U, Comey CT, Dirnhofer R. Typing of deoxyribonucleic acid (DNA) extracted from compact bone from human remains. J Forensic Sci. 1991;36(6):1649-61. [PubMed ID: 1685164].
-
5.
Lassen C, Hummel S, Herrmann B. Comparison of DNA extraction and amplification from ancient human bone and mummified soft tissue. Int J Legal Med. 1994;107(3):152-5. [PubMed ID: 7893611]. https://doi.org/10.1007/BF01225603.
-
6.
Ubelaker DH, Parra RC. Radiocarbon analysis of dental enamel and bone to evaluate date of birth and death: perspective from the southern hemisphere. Forensic Sci Int. 2011;208(1-3):103-7. [PubMed ID: 21167668]. https://doi.org/10.1016/j.forsciint.2010.11.013.
-
7.
Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2(7):1756-62. [PubMed ID: 17641642]. https://doi.org/10.1038/nprot.2007.247.
-
8.
von Wurmb-Schwark N, Heinrich A, Freudenberg M, Gebuhr M, Schwark T. The impact of DNA contamination of bone samples in forensic case analysis and anthropological research. Leg Med (Tokyo). 2008;10(3):125-30. [PubMed ID: 18035582]. https://doi.org/10.1016/j.legalmed.2007.10.001.
-
9.
Kemp BM, Smith DG. Use of bleach to eliminate contaminating DNA from the surface of bones and teeth. Forensic Sci Int. 2005;154(1):53-61. [PubMed ID: 16182949]. https://doi.org/10.1016/j.forsciint.2004.11.017.
-
10.
Liu Q, Liu L, Zhang M, Zhang Q, Wang Q, Ding X, et al. A Simple and Efficient Method of Extracting DNA from Aged Bones and Teeth. J Forensic Sci. 2018;63(3):824-8. [PubMed ID: 29240980]. https://doi.org/10.1111/1556-4029.13603.
-
11.
Loreille OM, Diegoli TM, Irwin JA, Coble MD, Parsons TJ. High efficiency DNA extraction from bone by total demineralization. Forensic Sci Int Genet. 2007;1(2):191-5. [PubMed ID: 19083754]. https://doi.org/10.1016/j.fsigen.2007.02.006.
-
12.
Holland MM, Cave CA, Holland CA, Bille TW. Development of a quality, high throughput DNA analysis procedure for skeletal samples to assist with the identification of victims from the World Trade Center attacks. Croat Med J. 2003;44(3):264-72. [PubMed ID: 12808717].
-
13.
Azlina A, Zurairah B, Ros SM, Idah MK, Rani SA. Extraction of mitochondrial DNA from tooth dentin: application of two techniques. Arch Orofacial Sci. 2011;6(1):9-14.
-
14.
Malaver PC, Yunis JJ. Different dental tissues as source of DNA for human identification in forensic cases. Croat Med J. 2003;44(3):306-9. [PubMed ID: 12808723].
-
15.
Pinchi V, Torricelli F, Nutini AL, Conti M, Iozzi S, Norelli GA. Techniques of dental DNA extraction: Some operative experiences. Forensic Sci Int. 2011;204(1-3):111-4. [PubMed ID: 20558019]. https://doi.org/10.1016/j.forsciint.2010.05.010.
-
16.
Cobb JC. Ancient DNA Recovered by a Non-destructive Method. Ancient Biomolecules. 2002;4(4):169-72. https://doi.org/10.1080/1358612021000028461.
-
17.
Smith BC, Fisher DL, Weedn VW, Warnock GR, Holland MM. A systematic approach to the sampling of dental DNA. J Forensic Sci. 1993;38(5):1194-209. [PubMed ID: 8228888].
-
18.
Marjanovic D, Durmic-Pasic A, Bakal N, Haveric S, Kalamujic B, Kovacevic L, et al. DNA identification of skeletal remains from the World War II mass graves uncovered in Slovenia. Croat Med J. 2007;48(4):513-9. [PubMed ID: 17696306]. [PubMed Central ID: PMC2080561].
-
19.
Presečki Ž, Brkić H, Primorac D, Drmić I. Methods of preparing the tooth for DNA isolation. Acta Stomatologica Croatica. 2000;34(1):21-4.
-
20.
Rubio L, Santos I, Gaitan MJ, Martin de-las Heras S. Time-dependent changes in DNA stability in decomposing teeth over 18 months. Acta Odontol Scand. 2013;71(3-4):638-43. [PubMed ID: 22783923]. https://doi.org/10.3109/00016357.2012.700068.
-
21.
Adler CJ, Haak W, Donlon D, Cooper A. Survival and recovery of DNA from ancient teeth and bones. J Archaeol Sci. 2011;38(5):956-64. https://doi.org/10.1016/j.jas.2010.11.010.
-
22.
Schwartz TR, Schwartz EA, Mieszerski L, McNally L, Kobilinsky L. Characterization of deoxyribonucleic acid (DNA) obtained from teeth subjected to various environmental conditions. J Forensic Sci. 1991;36(4):979-90. [PubMed ID: 1680960].
-
23.
Alvarez Garcia A, Munoz I, Pestoni C, Lareu MV, Rodriguez-Calvo MS, Carracedo A. Effect of environmental factors on PCR-DNA analysis from dental pulp. Int J Legal Med. 1996;109(3):125-9. [PubMed ID: 8956985]. https://doi.org/10.1007/BF01369671.
-
24.
Tilotta F, Brousseau P, Lepareur E, Yasukawa K, de Mazancourt P. A comparative study of two methods of dental pulp extraction for genetic fingerprinting. Forensic Sci Int. 2010;202(1-3):e39-43. [PubMed ID: 20663623]. https://doi.org/10.1016/j.forsciint.2010.06.019.
-
25.
Shiroma CY, Fielding CG, Lewis JJ, Gleisner MR, Dunn KN. A minimally destructive technique for sampling dentin powder for mitochondrial DNA testing. J Forensic Sci. 2004;49(4):791-5. [PubMed ID: 15317196].
-
26.
Palomo-Díez S, Martínez-Labarga C, Gomes C, Esparza-Arroyo Á, Rickards O, Arroyo-Pardo E. Comparison of two different DNA extraction methodologies for critical bone or teeth samples. Forensic Sci Int: Genet Suppl Series. 2017;6:e359-61. https://doi.org/10.1016/j.fsigss.2017.09.110.
-
27.
Mohammadi A, Alvanegh AG, Khafaei M, Azarian SH, Naderi M, Kiyani E, et al. A new and efficient method for DNA extraction from human skeletal remains usable in DNA typing. J Appl Biotechnol Rep. 2017;4(2):609-14.
-
28.
Bland JM, Altman DG. Measurement error proportional to the mean. BMJ. 1996;313(7049):106. [PubMed ID: 8688716]. [PubMed Central ID: PMC2351517]. https://doi.org/10.1136/bmj.313.7049.106.